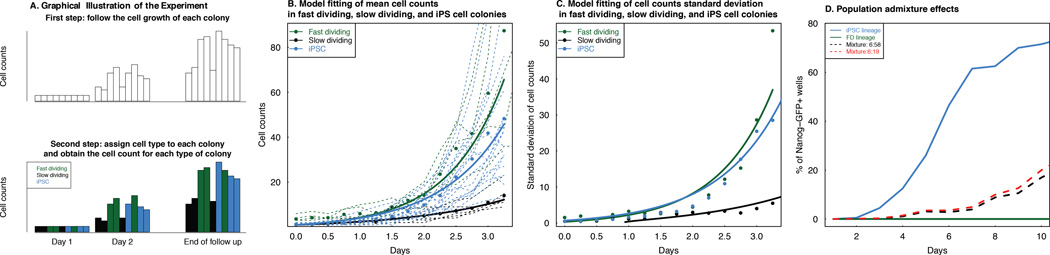
Figure 5. Validation of the model utility when cell count data is available.
A. A schematic description of a lineage tracing experiment (Smith et al. 2010) that assigned different morphological responses to OSKM induction in a standard reprogramming experiment using clonally inducible fibroblasts (fast dividing or FD, slow dividing or SD, and iPSC-generating, iPSC). Initially, labeled cells are tracked over time. Then, conditioning on colony formation or non-extinction, cell lineages are retrospectively assigned as FD (green), SD (grey), or iPSC (blue) and characterized as distinct groups. B. The mean cell count dynamics of FD, SD and iPSC are accurately described by our model. Since in the experiment no confluence was observed, the carrying capacity is set to infinity. The model prediction (lines) fit the observed cell counts very well (correlation above 0.95 in all three types of cells). Solid line: model-predicted cell counts over time; Dots: mean cell count dynamics averaging over all colonies belonging to each cell type; Dashed lines: cell counts for each colonies obtained from the data. C. The standard deviation of cell count dynamics of FD, SD and iPSC are also consistent with our model. Again the correlation between model prediction and data is above 0.95 in all three types of cells. Solid Line: model-predicted standard deviation of cell counts over time; Dots: standard deviation of cell counts obtained from the data. D. Population admixture of FD and iPSC cells can decrease the iPSC level dynamics compared to a homogeneous iPSC population. Blue solid line: uniform iPSC population; green solid line: uniform FD population; black dashed line: FD:iPSC = 6:58 mixture; red dashed line: FD:iPSC = 6:19 mixture.