Figure 2.
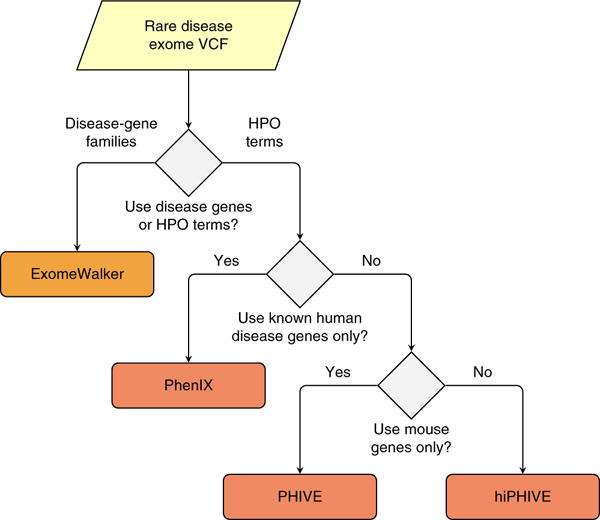
Choice of Exomiser prioritization method. Exomiser can be run using a set of seed genes if a disease-gene family can be identified. For instance, numerous genes mutated in retinitis pigmentosa (RP) have been identified. If it is suspected that a patient may have a mutation in a novel gene for RP, then Exomiser can be run with the ExomeWalker prioritization method together with a list of NCBI Entrez gene IDs for genes already known to be involved in RP (e.g., 6121 for RPE65, 130557 for ZNF513 and so on). Alternatively, Exomiser can be run using phenotypic similarity–based algorithms (PHIVE, PhenIX or hiPHIVE). Here the user needs to enter a list of HPO terms representing the clinical manifestations observed in the individual being investigated (Box 3). The choice of the prioritization method and parameters will depend on the clinical or research goal. PhenIX will interrogate only known Mendelian disease genes, and it will use only human phenotypic data to calculate phenotypic similarities. PHIVE will use mouse phenotypic data to identify candidates. Finally, hiPHIVE can use mouse, zebrafish and human clinical data, and it can additionally integrate further candidates on the basis of an analysis of the protein-protein interaction network. All of the analyses can be combined with additional filters (Box 1).
