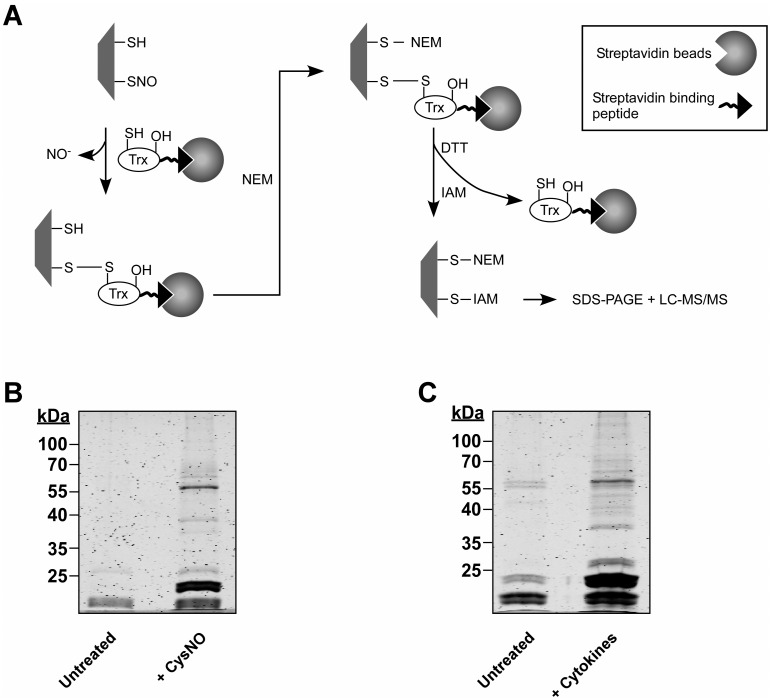
Fig 1. SNO trapping-based analysis of S-Nitrosylation in A549 cells.
(A) Schematic of the proteomic approach. Digitonin cell lysates, obtained from A549 treated with NO donor or with cytokines are incubated with a thioredoxin (Trx) trap mutant, Trx(C35S). In the trap mutant the resolving cysteine is replaced by serine (-OH). The protein also contains a streptavidin binding peptide. Trx(C35S) forms mixed disulfide bonds with nitrosylated substrates and the resulting complexes are pulled-down using avidin agarose. Identification of nitrosylation sites is assisted by differential thiol labeling, involving the sequential application of N-ethylmaleimide (NEM) and iodoacetamide (IAM). Proteins captured in the Trx pull-down are analyzed by SDS-PAGE or liquid chromatography-tandem mass spectrometry (LC-MS/MS). (B) A549 cells were treated with or without 500 μM S-nitrosocysteine (CysNO) for 10 min and thereafter digitonin lysates were incubated with Trx(C35S). Proteins captured by Trx were released by DTT and then analyzed by SDS-PAGE. Gels were stained with Krypton fluorescent protein stain and visualized using the Odyssey infrared imaging system. (C) A549 cells were treated for 72 h with LPS (0.5 μg/ml) and a cytokine mixture that included TNFα (20 ng/ml), IFN-γ (10 ng/ml) and IL-1β (10 ng/ml). Trx-based trapping of nitrosylated proteins was performed as in B.