Figure 2.
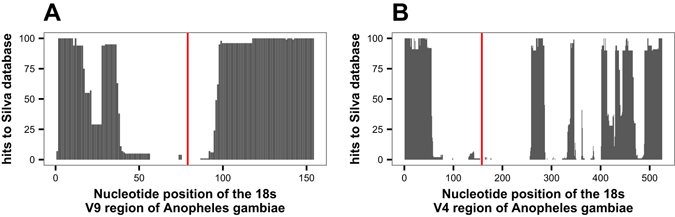
Identification of PNA oligonucleotide blocker candidates for the V9 region of Anopheles. In order to design PNA blockers specific to Anopheles, but allowing efficient amplification of other eukaryotic organisms, the Silva eukaryotic database was queried with mosquito candidate sequences to identify those with the smallest number of non-mosquito hits. (A) The full-length sequence of the V9 hypervariable region of A. gambiae was split in silico into all possible 17mers, and each 17mer was queried against the full length 18s rRNA gene sequences from the Silva 119 database for all non-mosquito eukaryotes. The number of matches was graphed (y-axis), limiting the representation to a maximum of 100 hits. The location of the selected current PNA blockers is indicated (red vertical line). (B) As in A, but for V4 region PNA oligonucleotide candidates.
