Figure 2.
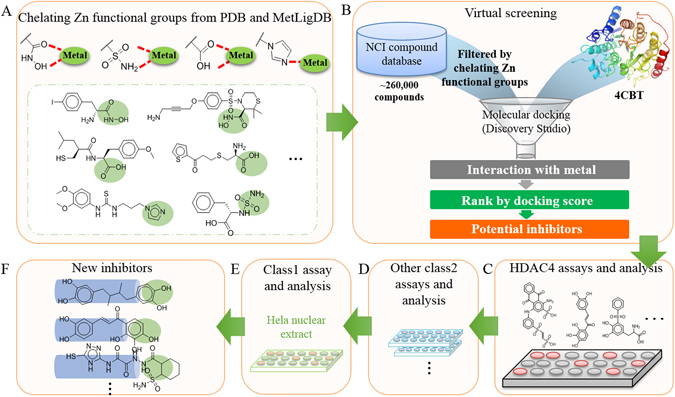
Framework of identifying non-hydroxamate inhibitors. (A) The structures of zinc binding compounds, which include hydroxamic acids, sulfonamides, carboxylic acids and imidazoles, are obtained from PDB and MetLigDB. The PDB and MetLigDB databases contains protein structures co-crystallized with metal binding ligands that can be used for novel inhibitor discovery (box with green dotted line, metal binding group located in green circle). (B) Structure-based virtual screening is performed by docking the NCI database compounds with the HDAC4 protein structure. The results are scored and ranked. The top 40 compounds were selected. (C) The candidate compounds undergo cellular assays to ensure docking results; the compounds with the strongest inhibitory effects were selected for further research. (D) The inhibitors were assayed against class IIa HDAC isozymes to determine inhibitory effects. (E) The inhibitors were assayed against class I HDAC to determine selectivity. (F) Discovered inhibitors with new ZBG that are selective for class IIa HDAC. Functional groups are indicated by green circle.
