Figure 2.
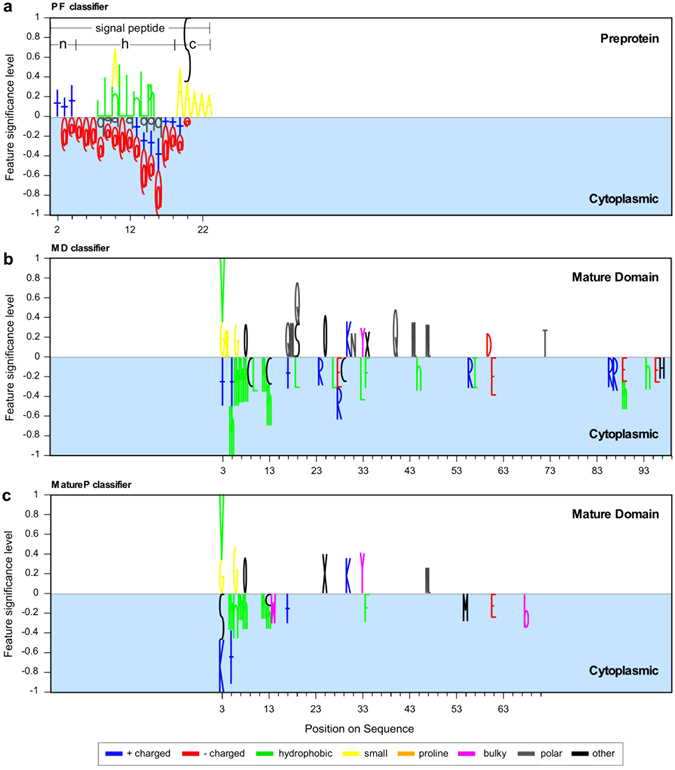
Representation of the selected features. Logo-like representation of the amino acid features taken into acount by each classifier: (a) the “preprotein”, (b) the “mature domain”, (c) and the MatureP. Different features at various positions on the protein sequence can be selected. The features correspond to either individual aminoacyl residues or groups of aminoacyl residues and are represented by a unique letter or symbol (see below). In (a) and (b) the complete set of the selected features is depicted whereas in (c) only the position specific amino acid features are applicable. The coefficients of the two linear classifiers, are the weights of the features which are employed here to represent the classifiers in Logo-like format (see Methods). If more than one features are selected at a position then a stack of symbols is drawn. The height of each stack is indicative of the significance of the position (see Methods). The weights have been normalized from −1 to 1 so that the classifiers are comparable (see Methods). Positively weighted features are selected for secretory whereas negatively for cytoplasmic proteins. In the “preprotein” classifier the most significant features are selected in the signal peptide region. However, there are also features selected in the mature domain region. When the signal peptide is removed (bottom) then more features are selected in the mature domains. A cluster of hydrophobic residues or arginine are disfavoured in the early mature domain (position 1 to 33). Symbols: @: (D,E); + : (K,R); sml: (V,G,A,P); sm: (A,G); h: (I,L,V,M); ph: (L,I,F); b: (Y,W,F); o- (T,S); x:(Y,T,S); pol: (N,Q,C); q: (N,Q,H).
