Figure 3.
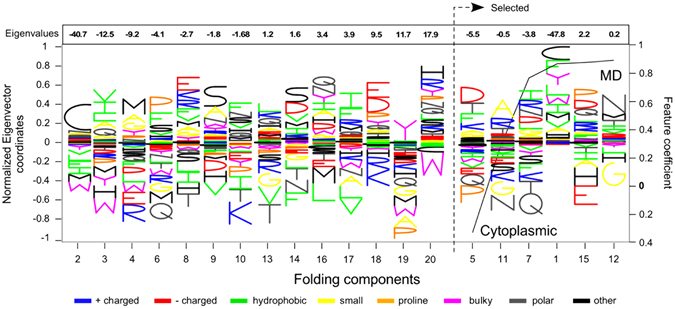
The selected folding features. Decomposition of the energy predictor matrix P to its eigenvectors37. Each element of the P matrix, tells how the energy of the residue of type i affects the occurrences of the residues of type j in the sequence37. We name the eigenvectors as folding components (FCs) because they represent a balance between stabilizing and destabilizing aminoacyl residues interactions. We represent each FC as a stack of letters (amino acids one letter code) where the size of each letter is proportional to the corresponding coordinate of the respective eigenvector. The black line denotes the weights of the linear classifier trained using the total energy per folding component (see Methods), termed as the “disorder classifier”. The FCs are rearranged per the weights of the classifier and are numbered in decreasing order of the respective eigenvalues, indicated at the top. Positively weighted FCs are selected for the mature domain sequences. For example, mature domains are composed mostly of Gln, Thr and Lys as the seventh FC dictates.
