Figure 3.
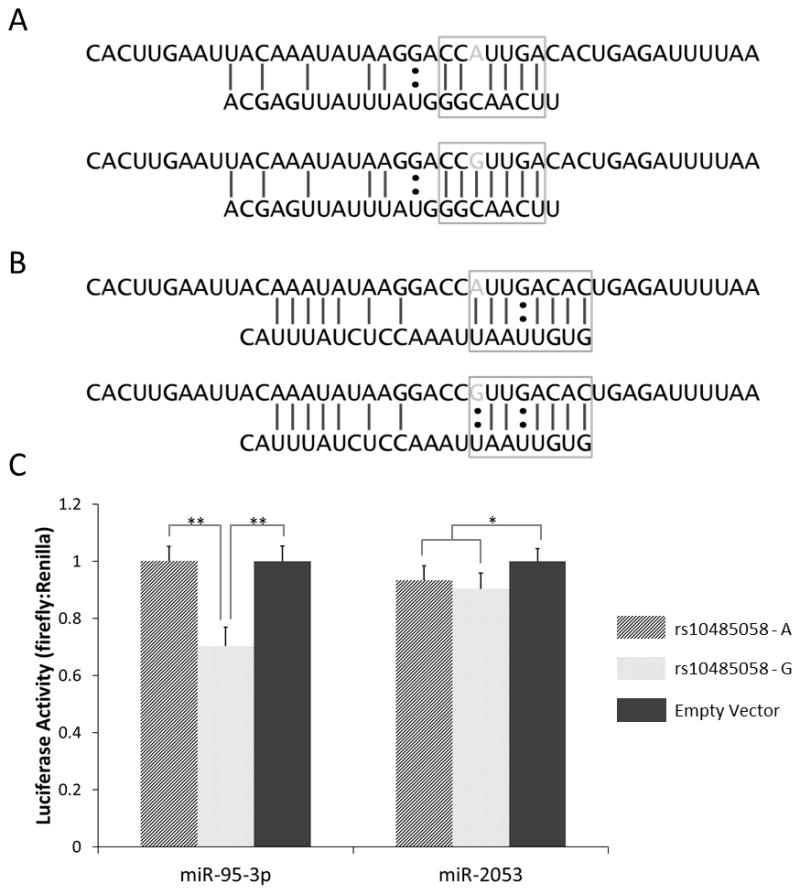
Predicted binding of miR-95-3p (A) and miR-2053 (B) to the genomic region containing the rs10485058 variant. RegRna (http://regrna.mbc.nctu.edu.tw/html/prediction.html) was used for in silico miRNA binding prediction. The top sequences represent mRNA, while the bottom sequences represent the respective miRNAs. The gray bases in the mRNA sequences indicate the rs10485058 allele. The seed sequence is boxed in each diagram. Perfect binding within the seed sequence area of miR-95-3p is predicted when the G allele is present, but not when the A allele is present. (C) Luciferase activity in BE(2)C cells co-transfected with mimics of miR-95-3p or miR-2053 and pmiR-Glo dual-luciferase constructs containing the rs10485058 locus in the 3′ UTR of firefly luciferase is shown. Luciferase activities were measured and the ratios of firefly:Renilla luciferase, normalized to the empty vector control, were graphed. Data are expressed as mean ± standard deviation (n = 9). * p < 0.05; ** p < 0.001
