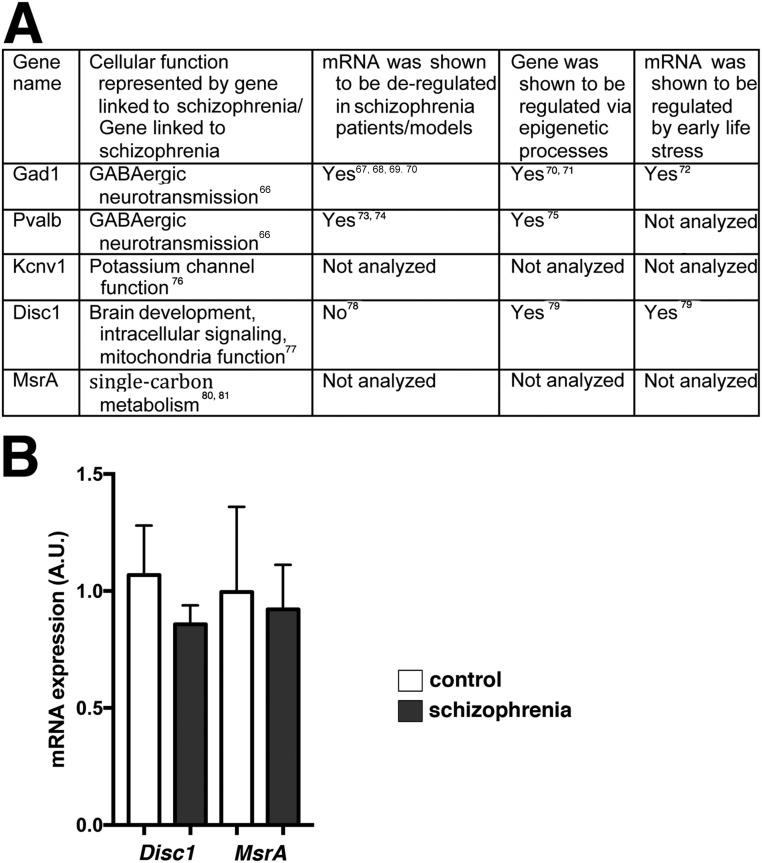
Fig. S8.
Selection of genes for HDAC1 ChIP analysis. (A) To test the impact of HDAC1 on gene expression directly, we decided to use an “educated guess” approach. Thus, we performed a literature search for genes that have been linked to schizophrenia or represent pathways linked to schizophrenia, were shown to be deregulated in patients with schizophrenia patients/models, and/or were affected by ELS. Our rational was to find genes down-regulated in the postmortem tissue samples from patients with schizophrenia who included in our study and that were also down-regulated in our experimental settings. We reasoned that such genes would be suitable examples to test the impact of HDAC1 on gene expression via ChIP analysis. On this basis, we initially selected five genes for qPCR analysis of postmortem tissue. In addition to Gad1, Pvalb, and Kcnv1, which are described in the main text (Fig. 4), we selected Disrupted in schizophrenia (Disc1) and methionine sulfoxide reductase A (MsrA), which have been linked to schizophrenia via genetic studies. (B) RT-qPCR analysis was performed using RNA from postmortem tissue (prefrontal cortex) of patients with schizophrenia and control individuals (same material as shown in Fig. 4). No significant difference in gene expression was observed for Disc1 and MsrA (Student’s t test). Both genes have been linked to schizophrenia via genetic studies. In addition, one study analyzed the expression of Disc1 in human brain tissue from patients with schizophrenia and found no difference (14). Our data confirm this observation. To the best of our knowledge, MsrA mRNA expression has not been tested before in brain tissue from patients with schizophrenia. Error bars indicate SEM.