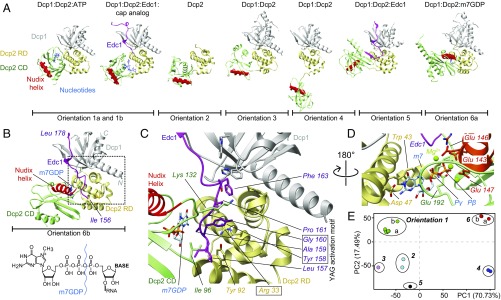
Fig. 1.
Static structures of the Dcp2 enzyme. (A) Known structures of Dcp2 display significantly different orientations of the Dcp2 RD (yellow) and CD (green). The catalytic Nudix helix in the CD is indicated in red, Dcp1 in gray, Edc1 in pink, and nucleotides in blue [orientation 1a, PDB ID code 2QKM (6); orientation 1b, 5KQ4 (11); orientation 2, 2A6T (12); orientation 3, 5LON (13); orientation 4, 2QKM (6); orientation 5, 5J3T (10); and orientation 6a, 5LOP (13)]. Note that the protein Edc3 that is present in the structure in orientation 6a is not shown for clarity. (B) Structure of the fully activated Dcp1:Dcp2:Edc1:m7GDP decapping complex (SI Appendix, Table S1). The YAG activation motif in Edc1 docks onto the C-terminal domain of Dcp2 and thereby locks Dcp2 such that the Nudix helix is in close proximity to the substrate binding site. The structure of the 5′ cap is shown and the bond that is hydrolyzed by Dcp2 is indicated. (C) Enlargement of the boxed region in B. Tyr-158 in the Edc1 YAG activation motif stacks between Arg-33 and Lys-132 in Dcp2. (D) The methylated base of the mRNA cap structure stacks onto Trp-43 in the Dcp2 RD and forms hydrogen bonds with Asp-47. Catalytically important Mg2+ ions are coordinated by Glu-143, 146, and 147 in the Dcp2 Nudix helix. (E) Based on a principle component analysis the known Dcp2 complexes can be separated into six groups.