Fig. 4.
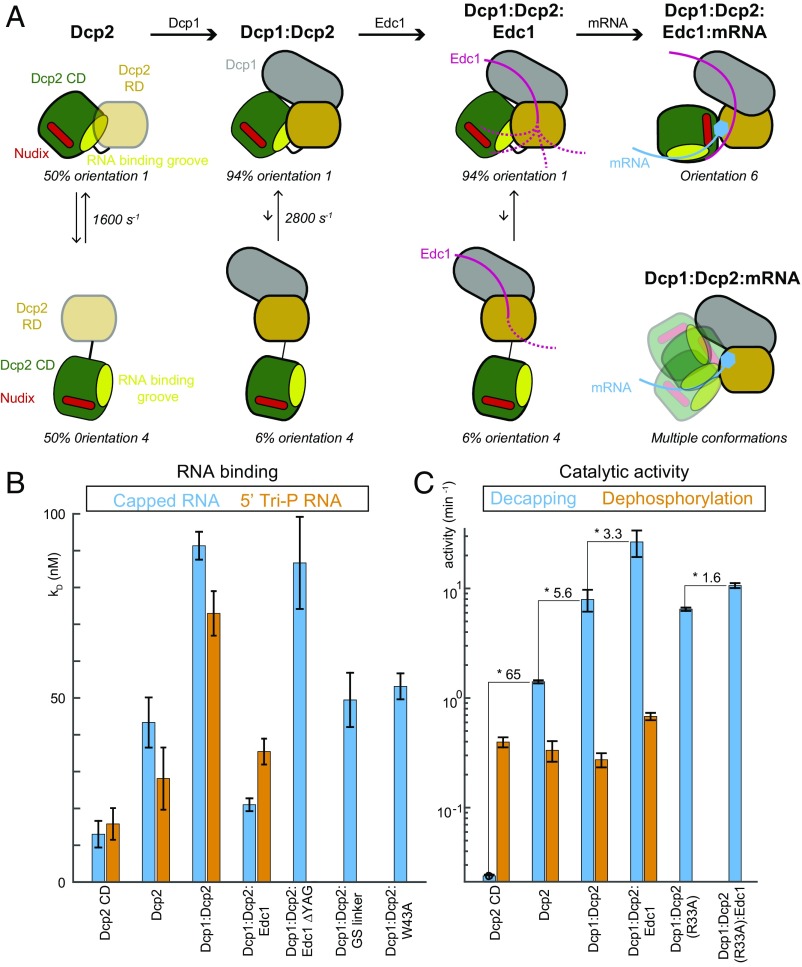
Summary of the conformational changes in Dcp2. (A) Cartoon representation of the Dcp2 domain orientations in the presence and absence of Dcp1, Edc1, and/ or substrate. In the apo Dcp2 enzyme the RD and CD domains exchange between an open (orientation 4) and a closed (orientation 1) state that are equally populated. Upon formation of the Dcp1:Dcp2 complex this equilibrium is shifted toward the closed state. Recruitment of Edc1 and the formation of the Dcp1:Dcp2:Edc1 complex has no effect on the Dcp2 domain orientations. In the Dcp1:Dcp2:Edc1:RNA complex the stable active Dcp2 conformation (orientation 6) is formed. In the absence of Edc1, the Dcp1:Dcp2:RNA complex is highly dynamic (Bottom Right). Substrate is recruited to the open form of these complexes, because there the RNA binding groove (light green) is fully accessible. The colors are as indicated in Fig. 1A; the low stability of the fold of the Dcp2 RD in the absence of Dcp1 is indicated with light orange. (B) Dissociation constants of a 30mer RNA for different Dcp2 complexes, where a higher bar indicates a weaker interaction. The RNA contained either a 5′ cap structure (blue) or a 5′ triphosphate (orange). The affinity decreases upon inclusion of the Dcp2 RD and Dcp1 and increases upon recruitment of Edc1. The Dcp1:Dcp2:Edc1 complex interacts most strongly with capped RNA. Removal of the YAG activation motif in Edc1 abolishes the effect of the activator. The extension of the linker between the Dcp2 RD and CD as well as the W43A mutation open the Dcp1:Dcp2 complex (Fig. 2E) and expose the RNA binding groove, thereby increasing the substrate affinity. (C) The activity of Dcp2 on capped RNA (blue, eukaryotic mRNA) and 5′ triphosphate RNA (orange, bacterial mRNA). The decapping activity (blue) increases from the Dcp2 CD toward the Dcp1:Dcp2:Edc1 complex. The increase in catalytic efficiency is largely abolished for a version of Dcp2 that cannot interact efficiently with the Edc1 YAG activation motif (Dcp2 R33A, Fig. 1C). The activity of Dcp2 on 5′ triphosphate RNA (orange) is largely independent of the Dcp2 RD, Dcp1, and Edc1.