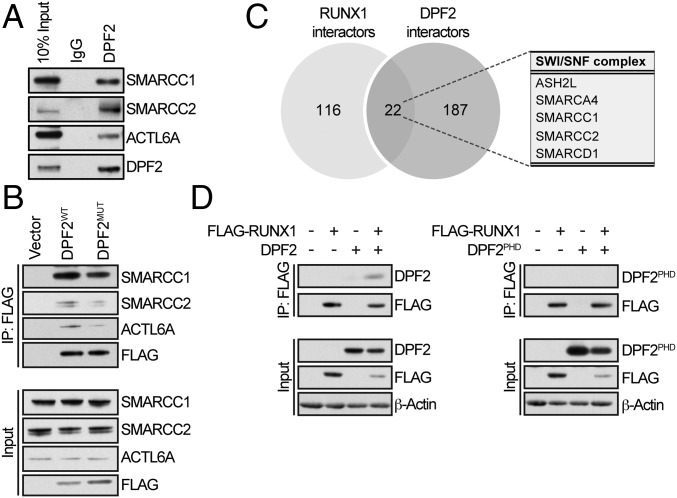
Fig. 5.
Protein–protein interaction network of DPF2. (A) Confirmation of SWI/SNF interacting proteins identified by the mass spectrometry analysis of proteins precipitated with endogenous DPF2. Whole-cell lysates from Kasumi-1 cells were immunoprecipitated with DPF2-conjugated beads and interacting proteins were analyzed by Western blot. (B) Confirmation of SWI/SNF interacting proteins identified by the mass spectrometry analysis using FLAG-DPF2 constructs. Wild-type FLAG-DPF2 (DPF2WT) and FLAG-DPF2MUT proteins were immunopreciptated from 293T cells using anti-FLAG magnetic beads and analyzed by Western blot. (C) Overlapping DPF2 and RUNX1 interaction partners. Comparison of mass spectrometry analysis of the proteins interacting with endogenous DPF2 and proteins interacting with the dimethyl-Arg223-RUNX1 peptide. DPF2 interacting proteins are normalized to an IgG control and methyl-RUNX1 interacting proteins are normalized to a nonmethylated peptide control. There were 22 protein interaction partners shared between the two datasets. (D) In vivo analysis of the DPF2–RUNX1 interaction. Coimmunoprecipitation of FLAG-RUNX1 with DPF2 and the isolated DPF2PHD domain. FLAG-RUNX1 and DPF2 or DPF2PHD were overexpressed in HEL cells by lentiviral transduction. Cells were lysed after 72 h, immunoprecipitated with anti-FLAG magnetic beads, and analyzed by Western blot. Input is 10% of the nuclear extract.