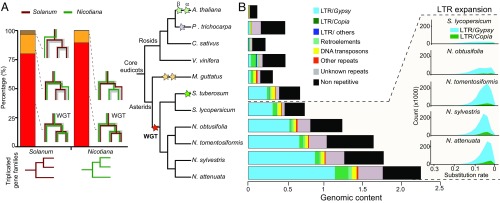
Fig. 1.
WGT in Nicotiana genomes is shared with other Solanaceae species but the Gypsy retrotransposons expansions are Nicotiana specific. Nicotiana genomes share the WGT with other Solanaceae species. (A, Left) Shared WGT event between Nicotiana and Solanum as revealed by superimposing species tree on the gene tree for the triplicated gene families in Nicotiana and Solanum. Red bar represents the percentage of triplication shared between Nicotiana and Solanum. Yellow bar represents shared duplication events or paralogous loss in one species. Brown bar represents Solanum-specific duplication events. (Right) Phylogenetic tree of 11 plant species and different colored stars indicate previously characterized whole-genome multiplication events. (B) Expansion of Gypsy transposable elements contributes substantially to genome size evolution in Nicotiana. (Left) Genomic content (1C content in gigabases) of repetitive versus nonrepetitive sequences in the 11 plant genomes. Black bar indicates nonrepetitive sequences, whereas other colors indicate repetitive sequences. (Right) Visualization of the expansion history of LTR retrotransposons in four Nicotiana genomes in comparison with tomato. The x axis (number of substitutions per site) refers to the divergence of a LTR from its closest paralog in the genome, with smaller numbers indicating more recent amplification events.