Fig. 5.
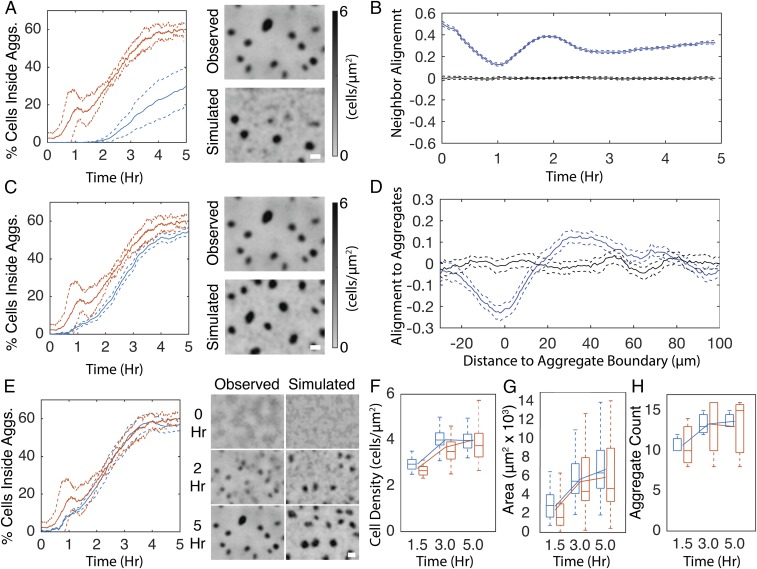
Closed-loop simulations reproduce wild-type–like aggregation with the addition of cell alignment. (A) Simulation results in which agents reduce movement proportional to cell density and perform a biased walk toward aggregates. (A, Left) Average (solid lines) and SD (dashed lines) of the percentage of cells inside detected aggregates for experimental (red) and simulation (blue) replicates. (A, Right) Comparison of the last frame of a representative experientially observed (Observed) cell density with a simulation (Simulated). (B) Average (solid lines) and 95% confidence intervals (dashed lines) of run vector alignment strength (blue lines) with neighboring run vectors that occurred within ±5 min and 15 μm. Black lines indicate alignment strength with randomly chosen runs. Values may span (−1,1) where 1 indicates all runs are parallel. Likewise, −1 indicates all runs are perpendicular. (C) Same as A with the addition that agents in the simulations align their orientation with neighboring agents. (D) Alignment strength of run vectors (blue lines) with vector pointing toward nearest aggregate centroid. Black lines indicate alignment strength after randomly shuffling each run’s distance to the nearest aggregate. Negative distances indicate that the run began inside an aggregate. Values may span (−1,1) as in B. (E–H) In addition to the agent behaviors from simulations in A and C, agents orient toward the nearest aggregate centroid. (E, Left) Percentage of cells inside aggregates as in A. (E, Right) Comparison of representative experientially observed cell density time progression with that observed in the closed-loop simulation. Grayscale is proportional to cell density as in A. (F) Average cell density inside aggregates. (G) Average aggregate area. (H) Aggregate count in each replicate. Box plots are formatted as in Fig. 1A. Lines indicate mean. (Scale bars, 100 μm.)