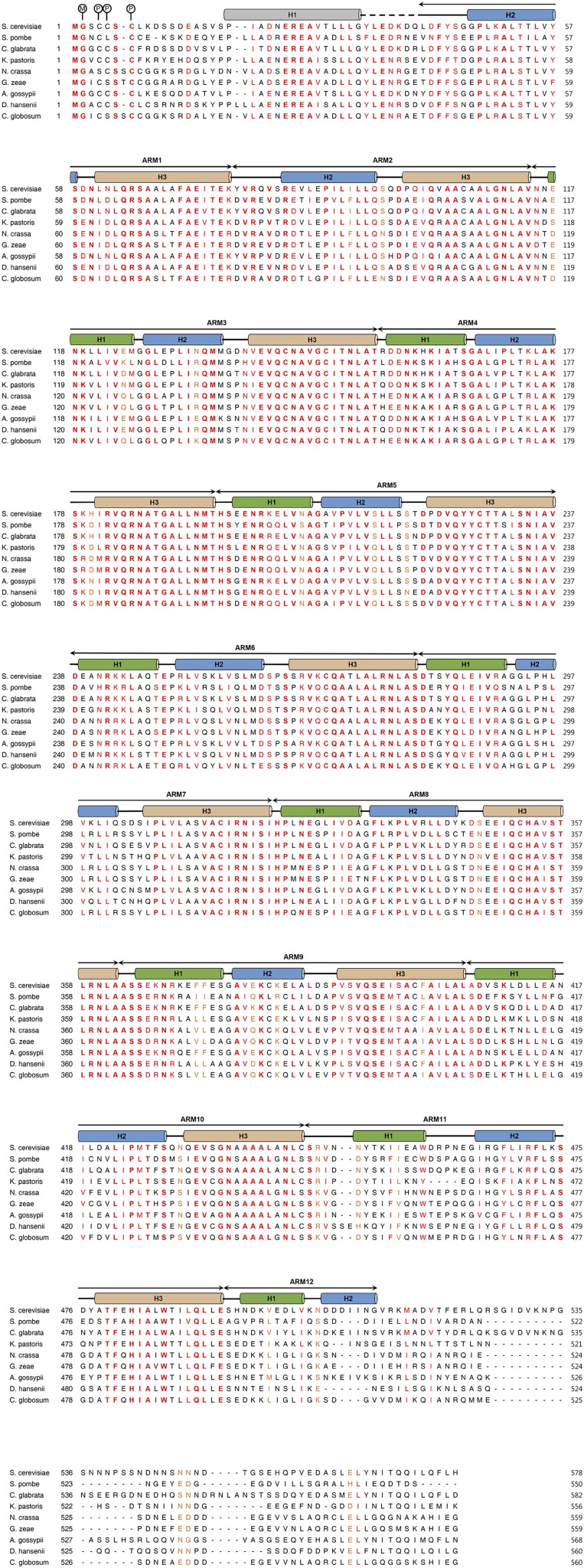
Fig. S1.
Sequence alignment of yeast Vac8p orthologs. Sequence alignment of full-length Vac8p orthologs in fungi. The N terminus of Vac8p is known to be modified by lipidation. The circled M and P indicate N-myristoyl glycine and S-palmitoyl cysteine, respectively. Secondary structural elements are indicated above the sequences with helices, β-stands, loops, and disordered regions depicted as cylinders, arrows, solid lines, and dashed lines, respectively. The crystal structure revealed that Vac8p is composed of an N-terminal helix (gray) followed by 12 ARMs. The three helices forming each ARM repeat are colored differently. Absolutely conserved, highly similar, and similar sequences are highlighted in bold red, red, and orange, respectively. UniProt entries for the sequences are as follows: S. cerevisiae (P39968), Schizosaccharomyces pombe (O43028), Candida glabrata (Q6FJV1), Komagataella pastoris (Q5EFZ4), Neurospora crassa (Q7RXW1), Gibberella zeae (Q4I1B1), Aphis gossypii (Q757R0), Debaryomyces hansenii (Q6BTZ4), and Chaetomium globosum (Q2GW27).