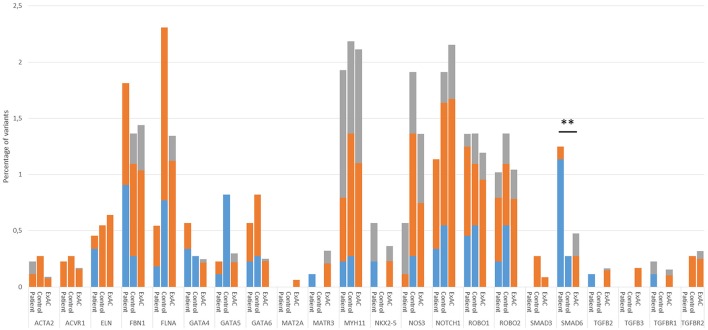
Figure 1.
Proportion of variant alleles per gene in the patient group, control group and ExAC cohort. Variants were selected as follows: First, we selected heterozygous coding or splice site variants with an allelic balance between 0.25 and 0.85 (FLNA in males: 0.75–1) and a minimum coverage of 10x. Next, we made three variant groups based on their frequency in the ExAC database; that is, variants that are absent from the ExAC control dataset (blue), variants with an ExAC MAF lower than 0.01% (orange) and variants with an ExAC MAF between 0.01% and 0.1% that had a CADD score above 20 (gray). Only statistics of the patient-ExAC comparison are shown (**p ≤ 0.01). No statistically significant differences in allele frequencies were observed between our control cohort and the ExAC controls. Abbreviations: ExAC, Exome Aggregation Consortium; MAF, Minor Allele frequency; CADD, Combined Annotation Dependent Depletion.