FIG 5.
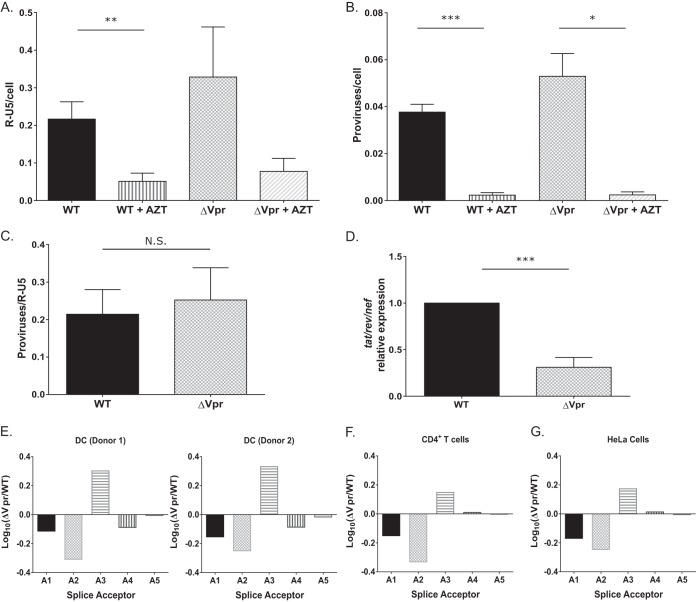
Viral transcription is attenuated in ΔVpr virus-infected MDDCs. (A to C) MDDCs infected with WT or ΔVpr viruses (MOI = 2) in the presence or absence of AZT (10 μM) were lysed at 72 h postinfection and processed for DNA isolation. Note that infected cells were cultured in the presence of indinavir (1 μM) to prevent viral spread. qPCR was used to detect early RT products (A) and integrated proviruses (B) by using the R-U5 and Alu-PCR primer sets, and the number of integrated proviruses was normalized to early RT products for each infection (C). The data reported are the means (± standard errors of the means) of results from three independent experiments. (D) The number of multiply spliced viral transcripts (tat-rev-nef) in MDDCs infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 1) was determined at 48 h postinfection by qRT-PCR. Viral transcripts were measured by using primers specific for tat-rev-nef multiply spliced transcripts. Data shown are means (± standard errors of the means) of results from four independent experiments with MDDCs derived from four donors. (E) Quantification of the 4-kb class of splice variants for MDDCs infected with Lai-YU2 or Lai-YU2/ΔVpr (MOI = 2) for 72 h. The data were normalized, log10 transformed, and then graphed according to slice acceptor usage. (E to G) Histograms show fold changes in splicing from D1 to each of the 5 viral splice acceptor sites A1 through A5 relative to a WT control. Splicing was quantified by using a Primer ID splicing assay for MDDCs from two independent infections (E) and productively infected CD4+ T cells (F) and HeLa cells (G), and data are from a single deep-sequencing experiment. Significance was calculated by using unpaired Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).