Abstract
Animals detect odorous chemicals through specialized olfactory sensory neurons (OSNs) that transduce odorants into neural electrical signals. We identified a novel and evolutionarily conserved protein, cilia- and flagella-associated protein 69 (CFAP69), in mice that regulates olfactory transduction kinetics. In the olfactory epithelium, CFAP69 is enriched in OSN cilia, where olfactory transduction occurs. Bioinformatic analysis suggests that a large portion of CFAP69 can form Armadillo-type α-helical repeats, which may mediate protein–protein interactions. OSNs lacking CFAP69, remarkably, displayed faster kinetics in both the on and off phases of electrophysiological responses at both the neuronal ensemble level as observed by electroolfactogram and the single-cell level as observed by single-cell suction pipette recordings. In single-cell analysis, OSNs lacking CFAP69 showed faster response integration and were able to fire APs more faithfully to repeated odor stimuli. Furthermore, both male and female mutant mice that specifically lack CFAP69 in OSNs exhibited attenuated performance in a buried food pellet test when a background of the same odor to the food pellet was present even though they should have better temporal resolution of coding olfactory stimulation at the peripheral. Therefore, the role of CFAP69 in the olfactory system seems to be to allow the olfactory transduction machinery to work at a precisely regulated range of response kinetics for robust olfactory behavior.
SIGNIFICANCE STATEMENT Sensory receptor cells are generally thought to evolve to respond to sensory cues as fast as they can. This idea is consistent with mutational analyses in various sensory systems, where mutations of sensory receptor cells often resulted in reduced response size and slowed response kinetics. Contrary to this idea, we have found that there is a kinetic “damper” present in the olfactory transduction cascade of the mouse that slows down the response kinetics and, by doing so, it reduces the peripheral temporal resolution in coding odor stimuli and allows for robust olfactory behavior. This study should trigger a rethinking of the significance of the intrinsic speed of sensory transduction and the pattern of the peripheral coding of sensory stimuli.
Keywords: CFAP69, olfaction
Introduction
Sensory receptor cells detect and transduce salient sensory stimuli into cellular electrical signals that encode the type, intensity, duration, and kinetics of the stimuli. These electrical signals are transmitted to and eventually interpreted by the brain to guide behavior. Although the mechanisms for sensory transduction vary among different systems, sensory receptor cells have evolved to be sensitive, rapid responding, and adaptable.
In the vertebrate olfactory system, olfactory sensory neurons (OSNs) in the nose detect and transduce odorous chemicals, or odorants, into membrane depolarization, which leads to generation and transmission of action potentials (APs) to the olfactory bulb of the brain (Firestein, 2001). Olfactory transduction takes place in the cilia of the OSN, which extend from the tip of the OSN dendrite into the mucus covering the nasal epithelium. In the vast majority of OSNs, transduction is mediated through a G-protein-coupled, cAMP-mediated signaling cascade (Kleene, 2008; Kaupp, 2010; Ferguson and Zhao, 2016). Specifically, the binding of an odorant to its G-protein-coupled odorant receptor (Buck and Axel, 1991) can lead to sequential activation of the odorant receptor, the olfactory G-protein Golf (Jones and Reed, 1989; Belluscio et al., 1998) and the G-protein effector adenylyl cyclase 3 (AC3) (Bakalyar and Reed, 1990; Wong et al., 2000). Activation of AC3 then leads to synthesis of cAMP, which in turn binds to and opens the olfactory cyclic-nucleotide-gated (CNG) channel (Dhallan et al., 1990; Brunet et al., 1996), allowing influx of cations Na+ and Ca2+ and triggering membrane depolarization. Intraciliary Ca2+ can open a calcium-activated chloride channel (Kleene and Gesteland, 1991; Kurahashi and Yau, 1993), Anoctamin 2 (ANO2) (Pifferi et al., 2009; Stephan et al., 2009; Rasche et al., 2010; Billig et al., 2011), which is responsible for Cl− efflux and further membrane depolarization. The electrical response of OSNs not only rapidly turns on, but also rapidly turns off when the odor stimulus is removed. Rapid termination of the response enables OSNs to recover sufficiently to respond to subsequent stimulation. To achieve rapid termination, OSNs actively remove cilial cAMP and Ca2+, thus closing the CNG channel and the chloride channel, respectively. The cAMP is degraded by phosphodiesterase 1 C in the cilia (Yan et al., 1995; Cygnar and Zhao, 2009) and Ca2+ is extruded from the cilia by a potassium-dependent sodium/calcium exchanger, NCKX4 (Stephan et al., 2011).
Despite substantial knowledge about the above-mentioned core components of the olfactory transduction cascade, less understood is how the transduction process is regulated to allow for proper sensitivity and response kinetics when responding to odors. To better understand olfactory transduction, we sought to investigate a novel protein, cilia- and flagella-associated protein 69 (CFAP69), which was found previously in an OSN cilial proteomic analysis (Stephan et al., 2009). The function of this protein had not been investigated in any system. By selectively knocking out the Cfap69 gene in mature OSNs, we were able to study the role of CFAP69 in olfaction.
Materials and Methods
Animals.
For all experiments involving mice, animals were handled and euthanized in accordance with methods approved by the Animal Care and Use Committees of each applicable institution. All analyses involving mice were performed on adult (2- to 8-month-old) mice. Experiments were performed on both male and female mice.
Evolutionary analysis.
The evolutionary history of the CFAP69 protein was inferred using the UPGMA method (Sneath and Sokal, 1973). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The analysis involved 10 aa sequences. All positions containing gaps and missing data were eliminated. There were a total of 683 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013).
Generation of conditional Cfap69 mutant mice.
The A330021E22Riktm1a(KOMP)Wtsi (abbreviated to Cfap69tm1a in this study) mouse strain was created from an embryonic stem cell clone (EPD0713_1_E05) generated by the Wellcome Trust Sanger Institute and made into mice by the KOMP Repository (www.KOMP.org) and the Mouse Biology Program (www.mousebiology.org) at the University of California–Davis. The Cfap69tm1a mice used in the present study were recovered from the cryopreserved embryos from KOMP by the Johns Hopkins University transgenic core facility. Cfap69tm1a mice carry a KO first allele in which a promoterless cassette including LacZ and neo genes were inserted in introns 4–5 of the Cfap69 gene. For the OSN-specific conditional KO mice, the Cfap69tm1a mice were crossed with the ubiquitously expressing Flippase line 129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J (The Jackson Laboratory) to excise the LacZ/neo cassette. These mice were then crossed with an OSN-specific Cre line B6;129P2-Omptm4(cre)Mom/MomJ (OmpCre) (The Jackson Laboratory) and then backcrossed for two to three generations to C57BL/6 mice to obtain offspring of several genotypes including the conditional CFAP69 mutant mice Cfap69flox/flox;OmpCre/+ and the control littermates Cfap69+/+;OmpCre/+. Primers TCAAACAGCACAGGAGATTCA (AT112) and TGCAAATGAATTAGCAGTATCTTCA (AT115), which span the floxed exon 5 region, were used to genotype the Cfap69 allele by PCR, with expected band sizes being 750 bp for WT and 921 bp for the floxed exon.
X-gal and immunofluorescent staining.
Deeply anesthetized mice (by Avertin) were transcardially perfused with PB, pH 7.4, followed by 4% (w/v) paraformaldehyde (4% PFA in PBS), and then postfixed in 4% PFA for 1 h. The tissue was decalcified in 500 mm EDTA in PBS for 1–2 d and then cryoprotected in 30% (w/v) sucrose in PBS for 1 d. The tissue was cut into 18-μm-thick coronal cryosections. For X-gal staining (Mombaerts et al., 1996), tissue sections were washed in PBS and incubated in PBS containing potassium ferricyanide, potassium ferrocyanide, and X-gal (Sigma-Aldrich) at 37°C for 12–16 h. For immunofluorescent staining, sections were incubated at 4°C overnight with primary antibodies (except for anti-CFAP69 antibody, which was incubated for 2 d) in PBS containing 0.1% (v/v) Triton X-100 and 1% (v/v) donkey serum. After washing, the sections were incubated with fluorescent secondary antibodies for 1 h at room temperature. After washing, the sections mounted in Fluoromount Aqueous Mounting Medium (Sigma-Aldrich) containing DAPI stain and imaged on an LSM 700 confocal microscope with Zen software (Zeiss). Primary antibodies were used at the following dilutions: anti-CFAP69 (rabbit, custom antibody), 1:100; anti-acetylated tubulin (mouse; Sigma-Aldrich T7451, RRID: AB_609894, 1:500; Ross et al., 2005; Tadenev et al., 2011); phalloidin–Alexa Fluor-488 (Thermo Fisher Scientific, A12379, 1:500); anti-Gγ13 (rabbit, 1:200, gift of R. Reed; Li et al., 2013), 1:200; anti-AC3 (rabbit, Santa Cruz Biotechnology sc-588, 1:200, RRID: AB_630839) (Zou et al., 2007); and anti-ANO2 (rabbit, Santa Cruz Biotechnology, sc-292004, RRID: AB_10844038, 1:100; Dibattista et al., 2012; Maurya and Menini, 2014). The following secondary antibodies were used in 1:400 dilutions: anti-rabbit–Alexa Fluor-488 (donkey, Thermo Fisher Scientific, A-21206) for anti-CFAP69, anti-Gγ13, and anti-ANO2; anti-rabbit–Alexa Fluor-546 (goat, Thermo Fisher Scientific, A-11029) for anti-AC3; and anti-mouse-Alexa-546 (goat, Thermo Fisher Scientific, A-21123) for anti-acetylated tubulin.
Custom antibodies.
Rabbit antibodies were generated against the antigenic peptide fragment CKVKPPLNDPKKSIPT, which spans aa 927–942 (the very C terminus) of the CFAP69 protein, by Thermo Fisher Scientific. Thermo Fisher Scientific performed the peptide synthesis, antibody generation, and affinity purification.
Cilia preparation.
A preparation enriched in olfactory cilia was prepared by the calcium shock method (Anholt et al., 1986). Briefly, deeply anesthetized mice were transcardially perfused with PBS to remove blood from the olfactory tissue. Olfactory mucosa were dissected into a solution containing the following (in mm): 120 NaCl, 5 KCl, 1.2 MgCl2, and 10 HEPES, pH 8.0, plus 10 mm CaCl2, and were treated by end-over-end rotation for 20 min at 4°C. The sample was centrifuged at low speed to pellet large cellular debris and the cilia in the supernatant were then transferred to a new tube. The cilia were pelleted under high-speed centrifugation (18,000 RCF) for 30 min and resuspended in a TEM buffer (10 mm Tris-HCl, 3 mm MgCl2, and 2 mm EDTA, pH 8.0).
Western blotting.
Olfactory epithelium (OE) tissues were homogenized and cilia preparations were dissolved in 2× Laemmli buffer followed by SDS-PAGE. After electrophoresis, the separated proteins were transferred onto a polyvinylidene difluoride membrane. The blot was blocked with 5% nonfat dry milk or 2% BSA and incubated overnight with primary antibodies at 4°C. After washing, the blot was incubated with HRP-linked secondary antibodies for 1 h at room temperature. After washing, the blot was treated with ECL-Plus reagent (Pierce) and exposed to film. Primary antibodies were used at the following dilutions: anti-CFAP69 (rabbit, custom antibody, 1:1000); anti-α-tubulin (mouse, Sigma-Aldrich T8203, RRID: AB_1841230), 1:10,000; anti-olfactory marker protein (OMP) (goat, Wako 544–10001, RRID: AB_664696, 1:10,000; Buiakova et al., 1996); and anti-AC3 (rabbit, Santa Cruz Biotechnology, SC-588, RRID: AB_630839, 1:1000; Zou et al., 2007). The following secondary antibodies were used at 1:2000 dilutions: anti-rabbit-HRP (goat, GE Healthcare, NA934) for anti-CFAP69 and anti-AC3; anti-goat-HRP (rabbit, Thermo Fisher Scientific, 61–1620) for anti-OMP; anti-mouse-HRP (sheep, GE Healthcare, NA931) for anti-α-tubulin.
Dolichos biflorus agglutinin (DBA) staining. Mice were deeply anesthetized by Avertin injection and decapitated. The head was bisected 1–2 mm off center and the septum dissected into 4% paraformaldehyde and fixed for 10 min at room temperature. Septa were rinsed 3× for 5 min in 1× PBS, blocked in 3% BSA in 1× PBS for 1 h at room temperature, and incubated with rhodamine-conjugated DBA (5 mg/ml, 1:500 dilution in blocking solution; Vector Laboratories) overnight at 4°C. Septa were then washed 3× for 5 min in 1× PBS. OE on both sides of the septum was peeled off into PBS, placed on a slide, mounted in Fluoromount Aqueous Mounting Medium, and imaged on an LSM 700 confocal microscope with Zen software (Zeiss). Only cells from the ventral region of the septum were examined. Cilia length was quantified in Fiji software (Schindelin et al., 2012) using the segmented line tool to trace cilia and the “measure” function to determine length. The mice used in this assay were from the Cfap69tm1b line, which was generated by crossing Cfap69tm1a to the early embryonically expressing Cre recombinase mouse line B6.FVB-Tg(EIIa-cre)C5379Lmgd/J (The Jackson Laboratory). Cfap69tm1b line is thus a Cfap69 whole-body KO line.
Cell proliferation assay.
Mice were injected intraperitoneally with 125 μg of 5-ethynyl-2′ deoxyuridine (EdU) (Thermo Fisher Scientific). Ten hours after the injection, the tissue was fixed and cryoprotected as described above. OE tissue was cut into 20 μm sections. EdU-labeled cells were detected using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific). Sections were mounted in Fluoromount-containing DAPI stain and imaged. The mice were 2 months old and were from 3 different litters.
EOG recordings.
EOG recordings were conducted as described previously (Cygnar et al., 2010). Amyl acetate and heptaldehyde were first diluted in DMSO to result in a series of stock solutions ranging from 5 × 10−6 m to 5 m, respectively. Each stock solution was then diluted 50-fold in water to generate a series of odorant solutions ranging from 1 × 10−7 to 0.1 m in concentrations. The 1 m amyl acetate solution was obtained by a 5-fold dilution of the 5 m stock in water. Vapor phase odorant was generated by putting 5 ml of an odorant solution of a given concentration in a sealed 60 ml glass bottle and letting the odorant solutions equilibrate in the bottles for at least 30 min. Delivery of odorant stimuli was controlled by a Picospritzer (Parker Hannifin). Note that the vapor concentration of odorants in each bottle is unknown, but will vary as a function of the concentration of odorants in the liquid phase. Even though the exact odorant concentrations are unknown, the odorant stimuli at the surface of the OE for a given concentration will be consistent between tissue preparations, allowing for comparison between WT and mutant mice. EOGs were recorded from a consistent position on turbinate IIB from the left half of the head. The data were collected and analyzed using AxoGraph Software (Molecular Devices) at a sampling rate of 1 kHz. All recordings were filtered at 25 Hz before analysis. For measuring termination time constants, the time windows used for the fit were as follows: 2.4–4 s for 10−6 m, 2.4–4.5 s for 10−5 m, 2.4–10 s for 10−4 m, 2.4–15 s for 10−3 m, and 2.4–20 s for 10−2–1 m.
OSN single-cell suction pipette recordings.
Mice were euthanized using CO2 followed by cervical dislocation. Single-cell suction recordings (Lowe and Gold, 1991; Reisert and Matthews, 1998) were performed as described previously (Ponissery Saidu et al., 2012). The cell body of an isolated OSN was sucked into the tip of a recording pipette, leaving the cilia and the dendritic knob accessible for solution changes. The recorded signals were sampled at 10 kHz using a Cambridge Electronic Design acquisition board and Signal software. Recordings were filtered at DC-50 Hz to monitor the receptor current and at DC-5000 Hz to also display the current for APs. All experiments were performed at 37°C. Rapid solution exchanges were achieved by transferring the tip of the recording pipette across the interface of neighboring streams of solution using the Perfusion Fast-Step SF-77B solution changer (Warner Instruments). Mammalian Ringer's solution contained the following (in mm): 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 0.01 EDTA, 10 HEPES, and 10 glucose. The pH was adjusted to 7.5 with NaOH. Odorant solutions were made daily from a stock containing 1 mm each of cineole and acetophenone. All chemicals were purchased from Sigma-Aldrich.
Buried food pellet test.
The buried food pellet test was performed using adult Cfap69+/+;OmpCre/+ (n = 22–23) and Cfap69flox/flox;OmpCre/+ (n = 22–25) mice at Zeitgeber time 8–1. All animals were housed individually with wood shavings as bedding for the duration of the experiment and had ad libitum access to water. Mice were weighed for 3 d before the start of the experiment to establish a baseline weight, deprived of food for 24 h before the experiment, and subsequently restricted to 0.12 g of rodent chow (Harlan Teklad) per gram body weight per day. Mice were weighed before the beginning of the experiment every day to make sure that they did not drop below 80% baseline body weight. The testing chambers were clean cages of dimensions 30 × 19 × 13 (L × W × H, in cm) filled with ∼650 cm3 of wood shavings as bedding. Two 40–60 mg pieces of de-creamed Oreo cookies (Nabisco) were buried just below the surface of the bedding in the following manner. The cage area was designated into halves length-wise. One cookie piece was buried in a randomized location within the left half, the other within the right half. Including two pellets in the experiment reduced the effective search area by half and better controlled for variance in the depth that the pellet was buried. In a single trial, a mouse was placed in the center of the cage and was given 200 s to locate either of the two pellets. Latency in finding the first pellet was recorded when the mouse touched the pellet. After the mouse located the first pellet, it was allowed to consume it. If a mouse failed to find a pellet within the allotted 200 s, the cookie pellet was exposed and presented to the mouse for subsequent consumption. After the trial, each mouse was returned to its respective cage. Mice were tested in a single trial per day for 10 consecutive days. On day 6, the pellet was positioned on the surface of the bedding for a visible pellet control trial. From day 7 on, mice were allotted 300 s to find the buried Oreo. On day 8, 1 g of powdered Oreo was infused evenly into the test bedding to produce background odor (low background). On day 9, 3 g of powdered Oreo was infused evenly into the test bedding (high background). On day 10, the bedding was once again free of background odor. The testing order of the animals was randomized for each day and fresh bedding was used every day for each mouse.
Statistical analyses.
Comparisons between two groups were determined by unpaired Student's t test or Fisher's exact test. Unless otherwise indicated, data are shown as mean ± SEM. Statistical difference was considered when p < 0.05.
Results
CFAP69 is a conserved protein enriched in OSN cilia
We became interested in CFAP69, which was originally annotated as Q8BH53, after it was found in an OSN cilial proteomic screen that was enriched for membrane proteins >55 kDa (Stephan et al., 2009). In addition to detecting known OSN cilial proteins such as AC3 and the CNG channel, the screen detected two proteins of unknown function, Q8BH53/CFAP69 and TMEM16B. TMEM16B, or Anoctamin 2 (ANO2), was shown to be a calcium-activated chloride channel (Pifferi et al., 2009; Stephan et al., 2009; Billig et al., 2011), but the function of CFAP69 remains unknown. Many other screens have also detected either Cfap69 (A330021E22Rik) transcript or CFAP69 protein in rat and mouse olfactory systems (Okazaki et al., 2002; Su et al., 2004; Sammeta et al., 2007; Mayer et al., 2009; Bennett et al., 2010; Rasche et al., 2010; Diez-Roux et al., 2011; Kanageswaran et al., 2015).
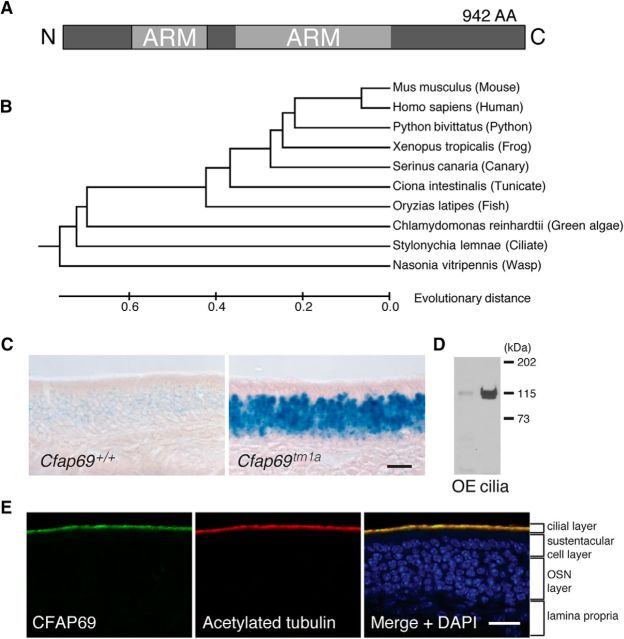
In mice, the Cfap69 gene is found on chromosome 5. The longest transcript is predicted to have 23 exons coding a protein of 942 aa (Ensembl ENSMUSG00000040473). A cDNA containing an open reading frame of 942 aa was amplified from the mouse nasal mucosa by RT-PCR. Based on bioinformatic analysis (InterProScan 5, Superfamily 1.75), a large portion of CFAP69 may form Armadillo-type α-helical repeats or ARM repeats (Fig. 1A; Gough et al., 2001; Jones et al., 2014). Although CFAP69 was originally annotated as a hypothetical transmembrane protein in the OSN cilial proteomic screen (Stephan et al., 2009), our hydropathy analysis (Mobyle@RPBS) failed to find any predicted transmembrane domains. Further bioinformatic analysis suggests that CFAP69 is evolutionally conserved among eukaryotes and can be found in humans, rodents, reptiles, amphibians, and some ciliated unicellular eukaryotes (Fig. 1B).
Figure 1.
CFAP69 is an evolutionarily conserved protein enriched in OSN cilia. A, Schematic of the CFAP69 protein. CFAP69 consists of 942 aa and is predicted to have ARM repeat domains. B, Evolutionary relationships of taxa based on protein sequences. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances. The evolutionary distances are in the units of the number of amino acid substitutions per site. The sum of branch length in the tree is 4.53259594. C, X-gal staining of OE sections from control and Cfap69tm1a mice. LacZ is expressed in the OE in Cfap69tm1a mice (right), but not in controls lacking the tm1a allele (left). Scale bar, 20 μm. D, Western blot analysis of OE and olfactory cilia preparations for CFAP69. Lanes were loaded with the same amount of total proteins. E, Immunofluorescent staining showing that CFAP69 is expressed in the cilial layer of the OE and colocalizes with acetylated tubulin, a marker of the cilia. Scale bar, 20 μm.
Using the Cfap69tm1a reporter mouse line (see Materials and Methods), we detected broad expression of the reporter gene LacZ in the OSN layer of the OE consistent with the only other report of Cfap69 expression (Fig. 1C; McClintock et al., 2008). Using antibodies against CFAP69, we detected a band of ∼115 kDa by Western blot analysis of OE and OE cilia preparations, which is in agreement with the calculated molecular weight of CFAP69 (106 kDa; Fig. 1D). In cilia preparation samples, a weaker band just below the 115 kDa band also appeared. This lower band perhaps corresponded to a smaller splice variant that we detected in the OE using 5′ rapid amplification of cDNA ends analysis. The shorter variant had an alternative transcription start site, but is in the same frame as the predominant 942 aa coding form and codes for a protein that is 897 aa. Immunostaining showed that CFAP69 is expressed in the cilial layer of the OE and colocalizes with a cilial marker, acetylated tubulin (Fig. 1E). Little CFAP69 expression was observed in the remainder of the tissue.
Knocking out Cfap69 in OSNs cause no overt structural and molecular alterations in the OE
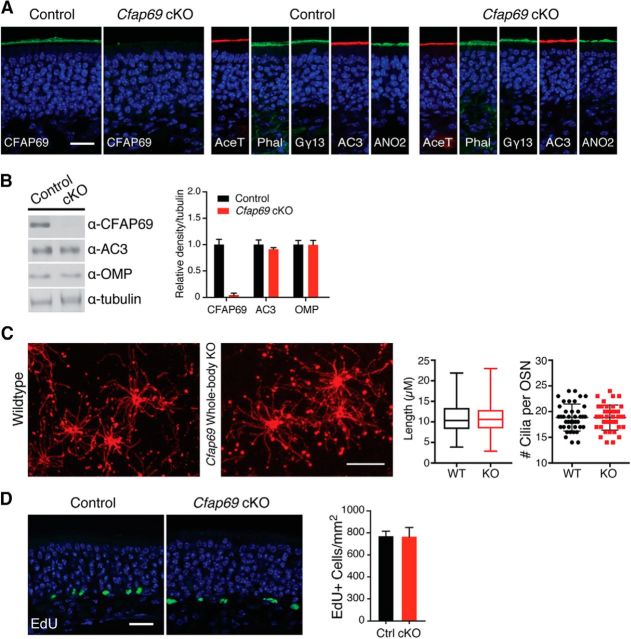
To study CFAP69 function in OSNs, we generated conditional Cfap69 KO mice (Cfap69flox/flox;OmpCre/+ mice, hereafter referred to as conditional Cfap69 mutants, or Cfap69 mutants) in which the Cfap69 gene is specifically knocked out in mature OSNs. We crossed the Cfap69tm1a line first to a ubiquitous-flippase mouse line to excise the LacZ/neo reporter cassette and generate the floxed allele of Cfap69. The Cfap69flox/flox mice were then crossed to mice carrying Cre-recombinase in the OMP locus. Conditional Cfap69 mutants showed no CFAP69 protein expression in the OE, as assayed by immunostaining and Western blotting (Fig. 2A,B).
Figure 2.
Conditional Cfap69 mutant mice have grossly normal OE. A, Immunofluorescent staining of OE sections from control and conditional Cfap69 mutant (Cfap69 cKO) mice. CFAP69 staining is absent in the conditional Cfap69 mutant mice. Staining of acetylated tubulin (AceT), Gγ13, AC3, ANO2, as well as phalloidin (Phol), which labels the apical microvilli of sustentacular cells, are comparable between the control and conditional Cfap69 mutant mice. Sections are counterstained with DAPI. Scale bar, 20 μm. B, Western blot analysis of total OE proteins. Right, Expression level relative to tubulin. Control, n = 4; Cfap69 cKO, n = 4 mice. Error bars indicate SEM. C, Left, Whole-mount preparation of septal OE with cilia of a subset of ventral region OSNs labeled by rhodamine-conjugated Dolichos biflorus agglutinin. Scale bar, 10 μm. Right, Quantification of the number of cilia per OSN (error bars indicate SEM) and their lengths. Box, Interquartile range; whiskers, minimum and maximum values. WT and Cfap69 whole-body KO, n = 4 mice with the cilia of 10 OSNs per animal examined. D, EdU labeling of proliferating cells in OE. Left, Proliferating cells are mostly found near the bottom of the OE in both genotypes. Sections are counterstained with DAPI. Scale bar, 20 μm. Right, Quantification. Control, n = 5; Cfap69 cKO n = 4 mice. Error bars indicate SEM.
Cfap69 mutants have no obvious abnormality in feeding and mating behaviors under the laboratory housing conditions. Mutants showed morphologically indistinguishable OE tissue compared with controls (Cfap69+/+;OmpCre/+ littermates, hereafter referred to as the control mice). Immunostaining against the cilial marker acetylated tubulin and the actin marker phalloidin, which stains microvilli of the supporting cells, appeared normal in the OE tissue (Fig. 2A). Typical expression and localization of olfactory transduction components, including Gγ13 (Li et al., 2013), AC3, and ANO2, was also observed (Fig. 2A). Western blot analysis showed no changes in the protein levels of AC3 and OMP, a mature OSN marker, in the OE tissue (Fig. 2B). In the whole-body KO mice, the number of cilia per OSN and the length of the cilia were not different from WT mice by DBA staining, which stains cilia of a subset of OSNs (Lipscomb et al., 2002; Challis et al., 2015; Fig. 2C). We also investigated cell proliferation in Cfap69 mutants and found no difference in the incorporation of the nucleotide analog EdU compared with the controls (Fig. 2D). Overall, we observed no overt structural or molecular alterations in the OE of conditional Cfap69 mutants.
Cfap69 mutant OSNs display faster response kinetics
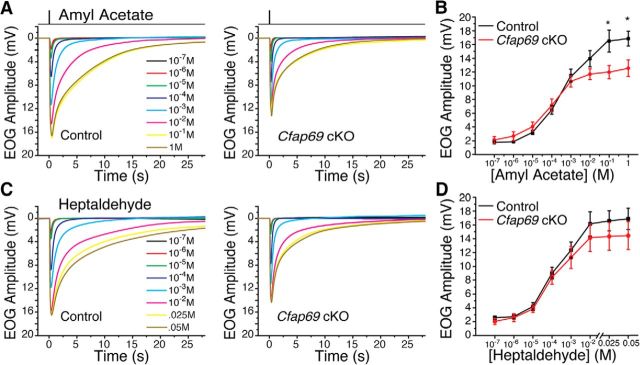
To investigate olfactory response in conditional Cfap69 mutants, we recorded EOG, the summed extracellular receptor potential from many OSNs measured at the OE surface (Scott and Scott-Johnson, 2002). Typically, a 100 ms odorant pulse elicits a dose-dependent response that peaks within 200–400 ms and decays thereafter in a few seconds. We found that conditional Cfap69 mutants displayed EOG amplitudes similar to the controls across a broad range of odorant concentrations from low to high when pulsed with two commonly used odorants, amyl acetate (Fig. 3A,B) and heptaldehyde (Fig. 3C,D). When stimulated with amyl acetate, however, the Cfap69 mutants did not reach the same maximal EOG amplitude at the highest odorant concentrations compared with the controls (Fig. 3A,B) and the response seemingly saturates at lower concentrations. When stimulated with heptaldehyde, the mutants showed similar responses throughout the experimental concentration range. The response amplitudes were reduced at the highest heptaldehyde concentrations, but the reductions were not statistically significant (Fig. 3C,D).
Figure 3.
CFAP69 mutant mice exhibit similar response size within a range of odorant concentrations: EOG analysis. A, B, EOG responses (A) and dose–response relationship (B) of peak EOG amplitudes evoked by 100 ms pulses of amyl acetate. Each trace in A represents the averaged EOG response across mice at the given concentration. Control, n = 8–12; Cfap69 cKO, n = 10–14 mice. Error bars indicate SEM. *p < 0.05, Student's t test. C, D, EOG responses (C) and dose–response relationship (D) of peak EOG amplitudes evoked by 100 ms pulses of heptaldehyde. Each trace in C represents the averaged EOG response across mice at the given concentration. Control, n = 7–10; Cfap69 cKO, n = 6–11 mice. Error bars indicate SEM, Student's t test. Note that the odorant concentrations indicated on the x-axis are the concentrations of the liquid solution from which the vapors of the odorant are generated.
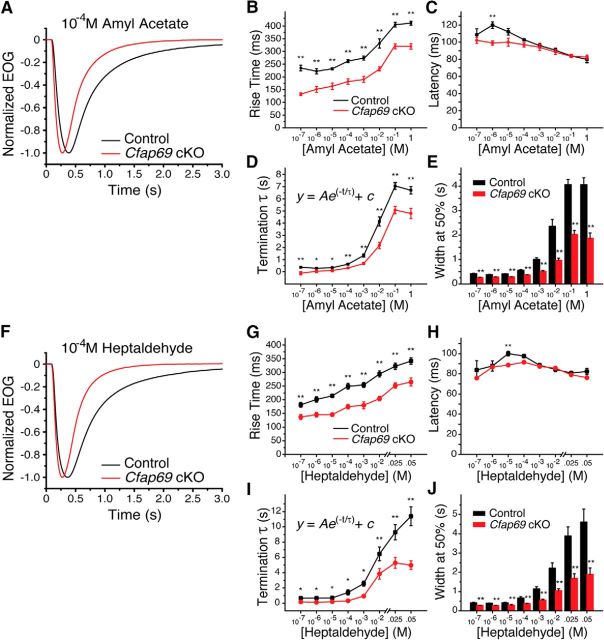
The most noticeable difference in the EOG between the control and the Cfap69 mutant was in the response kinetics. Cfap69 mutants displayed faster kinetics both in activation and termination of the response when pulsed with 100 ms amyl acetate or heptaldehyde (Fig. 4). Cfap69 mutants displayed a significantly faster rise time, measured as the time from 1% to 99% of the peak, across all odorant concentrations for the two odorants tested (Fig. 4B,G). The activation latency, defined as the time from the start of odorant stimulation to 1% of the peak, was comparable between control mice and Cfap69 mutants at most of the experimental concentrations (Fig. 4C,H), whereas, at one low concentration of each odorant (10−6 m amyl acetate or 10−5 m heptaldehyde), Cfap69 mutants displayed decreased latency compared with the controls. Cfap69 mutants also displayed faster termination of the response. The response termination rate was measured by the time constant (τ), which was obtained by fitting the EOG termination phase with a single exponential decay. Cfap69 mutants had significantly deceased time constant across all odorant concentrations for the two odorants tested (Fig. 4D,I). The faster kinetics both in activation and termination resulted in more transient EOG responses in Cfap69 mutants than in controls, as measured by the duration between the two time points when the EOG amplitudes are half of the peak (Fig. 4E,J).
Figure 4.
CFAP69 slows down OSN response kinetics: EOG analysis. A–E, Responses to 100 ms pulses of amyl acetate. A, Amplitude-normalized EOG responses to 10−4 m amyl acetate. Each trace represents the averaged EOG response across mice. Control, n = 11, Cfap69 cKO, n = 10 mice. B, EOG rise time, the time from 1% to 99% of the peak amplitude. C, EOG activation latency, the time from stimulation onset to 1% of the peak amplitude. D, EOG termination rate. The time constant (τ) is determined by fitting a single exponential function to the termination phase of the EOG trace. E, Width (timespan) of the EOG response at 50% of the peak amplitude. In B–E, Control, n = 8–12; Cfap69 cKO, n = 10–14 mice. *p < 0.05; **p < 0.01. Error bars indicate SEM. Student's t test. F–J, Responses to 100 ms pulses of heptaldehyde. F, Amplitude-normalized EOG responses to 10−4 m heptaldehyde. Each trace represents the averaged EOG response across mice. Control, n = 10; Cfap69 cKO, n = 10 mice. G, EOG rise time. H, EOG activation latency. I, EOG termination rate. J, Width (timespan) of the EOG response at 50% of the peak amplitude. G–J, Control, n = 7–10; Cfap69 cKO, n = 6–11 mice. *p < 0.05; **p < 0.01. Error bars indicate SEM, Student's t test. Note that the odorant concentrations indicated on the x-axis are the concentrations of the liquid solution from which the vapors of the odorant are generated.
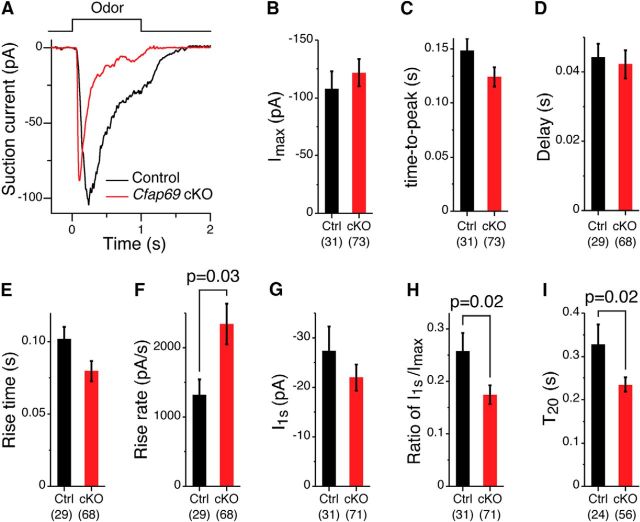
We next investigated how odor response might be altered at the single-cell level using suction pipette recordings. In this experiment, suction current responses of individual OSNs to odorant pulses were recorded from OSN cell bodies and an odorant pulse was applied at the exposed cilia and dendritic knob (Ponissery Saidu et al., 2012). Typically, a responsive OSN generates a quickly increasing current after a short delay; the current declines to a lower level after reaching its peak and returns back to baseline after odorant exposure ceases. The suction pipette recording technique also allows for monitoring the generation of APs. OSNs were exposed for 1 s (-sec) to the odorant mixture (100 μm each of cineole and acetophenone; Fig. 5A). OSNs lacking CFAP69 generated receptor currents with response magnitude (Imax) comparable to that of the control OSNs (Fig. 5B). The Cfap69 mutant OSNs and the control OSNs showed no significant difference in parameters including time-to-peak (Fig. 5C), the time between stimulation onset and the peak current; the response delay (Fig. 5D), measured as the time between stimulation onset and the generation of the first AP; and the rise time (Fig. 5E), measured as the time-to-peak minus the response delay. The mutant OSNs, however, showed a larger rise rate of the receptor current, which is measured as the rise time divided by Imax (Fig. 5F). During the 1 s stimulation, the receptor current of mutant OSNs declined more than that of control OSNs, as manifested by a smaller ratio of current at the end of 1 s stimulation (I1s)/Imax in the mutant OSNs (Fig. 5G,H). The response termination rate was measured by T20, which is the time required for the current at 1 s to fall to 20% of its value. The T20 of the mutant OSN was significantly smaller than that of the control OSN (Fig. 5I). Here, the single-cell data show changes in response kinetics in Cfap69 mutant OSNs, including faster rise rate, more transient response, and faster termination, consistent with the EOG results. Together, these data suggest that CFAP69 functions to slow down both the activation and the shutoff of the odor response in OSNs.
Figure 5.
CFAP69 slows down OSN response kinetics: single-cell analysis. A, Representative suction current traces of control and Cfap69 cKO OSNs to a 1 s pulse of 100 μm each of cineole and acetophenone. B, Imax, the peak amplitude of the suction current. C, Time-to-peak, the time from stimulation onset to the peak amplitude of the current. D, Response delay, the time between stimulation onset and the first AP. E, Rise time, measured as the time-to-peak minus the response delay. F, Rise rate, measured as Imax divided by the rise time. G, I1s, the current at the end of 1 s stimulation. H, I1s/Imax. I, T20, the time for the response to fall to 20% of I1s. In B–I, OSN numbers are shown in the parentheses. Error bars indicate SEM, Student's t test.
Cfap69 mutant OSNs integrate the stimulus faster
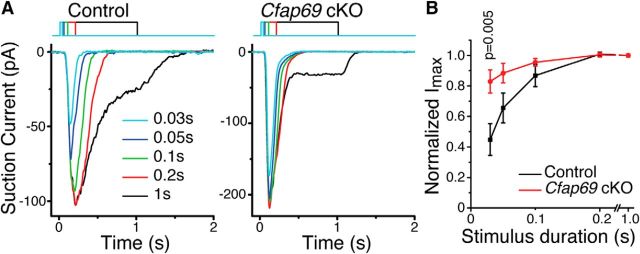
OSNs integrate the odor stimulus over time (Firestein et al., 1990; Firestein et al., 1993; Bhandawat et al., 2005) such that longer exposures yield larger responses. In single-cell experiments, we delivered odorant pulses (100 μm each of cineole and acetophenone) of increasing duration to determine whether response integration was altered in OSNs lacking CFAP69. A 30 ms pulse to odorants induced receptor currents that were 45% of the maximum in control OSNs; the response increased with increasing stimulus duration until the stimulus duration was ∼0.2 s. Further prolonging the stimulation did not further increase the response amplitude (Fig. 6A,B). In contrast, even at the shortest (30 ms) stimulus duration, OSNs lacking CFAP69 already reached ∼82% of their maximal response (Fig. 6B). Therefore, OSNs lacking CFAP69 integrate the stimulus faster.
Figure 6.
CFAP69 slows down OSN response integration. A, Representative traces from control and Cfap69 cKO OSNs when exposed to varying lengths of odorant stimulation. B, Normalized peak current (Imax). Control, n = 13; Cfap69 cKO, n = 16 OSNs. Error bars indicate SEM, Student's t test.
Cfap69 mutant OSNs fire APs more faithfully to repeated stimuli
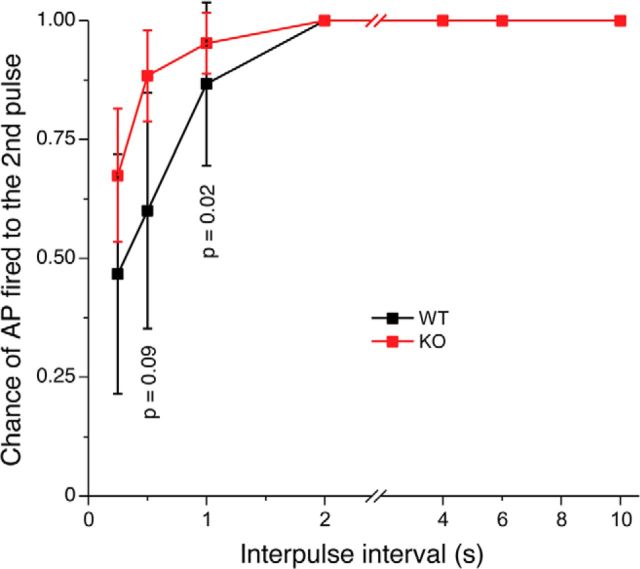
APs are typically only generated during the activation phase of the receptor current (Reisert and Matthews, 2001; Ghatpande and Reisert, 2011). Because Cfap69 mutant OSNs exhibited faster response kinetics, we investigated how such faster kinetics might affect the encoding of APs. We delivered 2 1 s odorant pulses (100 μm each of cineole and acetophenone) with varied interpulse, or recovery, interval to individual OSNs. The interpulse interval varied from 0.25 to 10 s. Both the control and Cfap69 mutant OSNs generated APs 100% of the time to the first pulse. We then examined the chance that an OSN generated an AP in response to the second pulse. When the interpulse interval is short, OSNs often fail to generate APs to the second pulse if the response to the first has not yet terminated. Under such conditions, OSNs are still sufficiently depolarized to maintain voltage-gated Na+ channel inactivation (Trotier and MacLeod, 1983; Trotier, 1994; Reisert and Matthews, 1999, 2001) and prevent AP generation by the second pulse. We found that the chance of a Cfap69 mutant OSN being able to generate APs to the second pulse was significantly greater than control OSNs at short interpulse intervals (Fig. 7). After a 0.5 s interpulse period, Cfap69 mutant OSNs generated an AP at 89% of the time, whereas controls OSNs only generated an AP at 60% of the time (Fig. 7). Therefore, Cfap69 mutant OSNs fire APs to repeated stimuli more faithfully than control OSNs. The control and Cfap69 OSNs only reliably generated an AP when the interpulse interval was ≥2 s. These data suggest that CFAP69, by slowing down the transduction kinetics, reduces temporal resolution and reliability of AP generation of OSNs in coding odor stimuli.
Figure 7.
Cfap69 mutant OSNs fire APs to repeated stimuli more faithfully than control OSNs. Individual OSNs were stimulated with two pulses of an odorant mixture with varied interpulse interval. The chance of an AP is generated to the second pulse is plotted against the interpulse interval time. Control, n = 14–15; Cfap69 cKO, n = 35–43 OSNs. Error bars indicate ± 95% confidence intervals, Fisher's exact test.
Cfap69 mutant mice show attenuated performance in an olfactory behavioral task
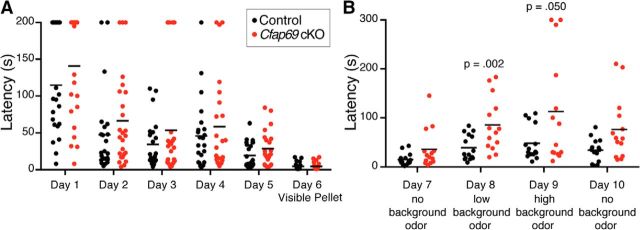
Given that Cfap69 mutant OSNs exhibited alterations in the way that they relay odor information, we examined how such alterations may influence odor perception in a behavioral assay. We chose the buried food pellet test, in which food-restricted mice need to use their olfactory sense to locate a buried food pellet under bedding (Stephan et al., 2011; Pietra et al., 2016). Over the course of the first phase of the experiment, both the control mice and Cfap69 conditional mutants were able to locate the buried food pellet with increasing rapidity, measured as the time to reach the pellet, over 5 d (Fig. 8A). Cfap69 mutants took slightly but not significantly longer time to locate the pellet. To control for any motivational, cognitive, or motor defects, the pellet was positioned on the surface of the bedding on day 6 (visible pellet). Both the control and Cfap69 mutant mice were able to locate the pellet equally rapidly (Fig. 8A).
Figure 8.
Conditional Cfap69 mutant mice show attenuated performance in an odor-localization behavioral task. A, Time to reach the first food pellet in the buried food pellet test. The time to locate the food pellet is plotted against the trial day for each individual mouse. On day 6 (visible pellet), the pellet was left on the surface of the bedding to be visible to the mouse. Controls, n = 22–23; Cfap69 cKO, n = 22–25 mice, Student's t test. B, Time to reach the first pellet when a background odor that is the same as the food pellet is present. Controls, n = 14–15; Cfap69 cKO, n = 14 mice, Student's t test.
In the second phase of the experiment, we infused powdered food into the bedding to create a background of the same odor. On day 7, the mice performed the same task as in days 1–5 and there was no background food odor. On this day, both the control and Cfap69 mutant mice were able to locate the pellet in a similar amount of time (Fig. 8B). On day 8 (“low background odor”), a small amount of powdered food was added to the bedding in the test cage. Control mice took a slightly but not significantly longer time to locate the pellet compared with the previous day. However, conditional Cfap69 mutants took significantly longer to locate the buried food pellet than the control animals did. On day 9 (“high background odor”), a larger amount of powdered food was infused into the bedding. Again, control mice performed this task comparably to day 8. Cfap69 conditional mutants took even more time to locate the buried food pellet (Fig. 8B). On day 10, when there was again no background odor, Cfap69 conditional mutants then took longer, but not significantly longer, to locate the pellet (Fig. 8B). These behavioral assays suggest that CFAP69 is required for challenging olfactory tasks.
Discussion
In this study, we identify a novel protein, CFAP69, in mice that is enriched in olfactory cilia and plays a critical role in regulating the response of OSNs, especially the response kinetics. OSNs lacking CFAP69 displayed faster kinetics in both onset and offset of electrophysiological responses and were able to fire APs more faithfully to repeated stimuli. In mammalian OSNs, aside from the core transduction components, several proteins, including OMP (Buiakova et al., 1996; Ivic et al., 2000; Reisert et al., 2007; Lee et al., 2011), Ric-8b (Von Dannecker et al., 2005; Kerr et al., 2008), RGS2 (Sinnarajah et al., 2001), MUPP1 (Dooley et al., 2009; Baumgart et al., 2014), and Goofy (Kaneko-Goto et al., 2013), have been shown previously to regulate olfactory signaling. All of these proteins, except RGS2, perform functions enhancing the transduction process. Although blockage of RGS2 leads to increased electrophysiological response (Sinnarajah et al., 2001), no protein has been shown previously to slow down OSN response kinetics. CFAP69 thus represents a new and unconventional regulator of the olfactory transduction process.
In sensory biology, it is seemingly reasonable that sensory receptor cells evolve to respond to sensory cues as fast as possible. This thought is consistent with mutational analyses in various sensory systems, where mutations of sensory receptor cells often resulted in reduced sensitivity and slowed response kinetics. From a systems perspective, it is also conceivable that a sensor should function as quickly as it can to report the information of stimuli, both on and off, to the next processing stage and a fast response kinetics is essential for such purpose. Contrary to this thought, we have found that there is a native “damper” present to slow down a sensory transduction process and, by doing so, it reduces the peripheral temporal resolution in coding sensory stimuli.
An insightful finding of this study is that, even though Cfap69 mutant mice should have better temporal resolution of coding olfactory stimuli at the peripheral sensory receptor level, they performed inferiorly in an olfactory behavioral task under certain conditions. Under the conditions used in the buried food pellet test, the loss of CFAP69 in OSNs led to poorer performance of the animal when a background of the same odor to the food pellet was present, although the loss of CFAP69 is tolerable when such background odor was not applied. These behavioral assays suggest that CFAP69 may be more needed for food-finding behaviors as occur in natural settings, where many salient background odors are present. Therefore, faster transduction kinetics and better temporal resolution in coding olfactory stimuli at the peripheral sensory receptor cell level is not always beneficial to the evolutionary fitness of the animal.
How could faster transduction kinetics lead to a reduced ability of mice to find an odor source in the presence of a background of the same odor? The faster transduction kinetics caused by the lack of CFAP69 apparently affects the AP coding property of OSNs. The fact that Cfap69 mutant OSNs can fire APs more faithfully to a recurring stimulus (Fig. 7) despite the adaptation effect (Fig. 5H) caused by the 1 s sustained exposure is consistent with the idea that the adaptive filter (Ghatpande and Reisert, 2011) of Cfap69 OSNs in coding odor stimuli is impaired. A more faithful ability in generating APs in response to repeated stimuli indicates that mutant OSNs extend their ability in coding odor stimuli to higher stimulation frequencies. This extended bandwidth, likely due to impairment of the adaptive filter, would result in extended and increased signal input to the olfactory bulb. This also potentially increases the noise input to the olfactory bulb. When a background of the same odor is present, the central olfactory circuit of Cfap69 mutant mice for detecting this odor could be overwhelmed by a barrage of signal inputs plus associated noise from OSNs due to the extended bandwidth, explaining the observed behavioral deficit. Such alteration in the coding property of OSNs seems to be tolerable in “simple” situations such as in the absence of the background odor, when the amount of input from OSNs is limited. In addition, when responding to an odorant stimulus, OSNs likely code the “molecular flux” instead of simply the odorant concentration at the level of transduction (Firestein and Shepherd, 1991). The faster stimulus integration of mutant OSNs (Fig. 6) means that mutant OSNs have a shorter time to estimate an odorant concentration. When a background of the same odor as that of the food pellet was present in the buried food pellet test, the mice needed to detect an odor against a background of lower concentration. It is also possible that the behavioral deficit of mutant mice stems from the altered integration property of OSNs, which affords shorter time periods for mutant mice at the behavioral level to detect concentration differences. Regardless, CFAP69 allows the olfactory transduction machinery to work at a properly regulated range of response kinetics for robust olfactory behavior.
We found that Cfap69 mutant OSNs displayed similar response size to the control OSNs in most of our electrophysiological recordings. In the single-cell analysis, Cfap69 mutant OSNs and the control OSNs showed similar response sizes to the mix of cineole and acetophenone at the concentration used. In the EOG analysis, we did observe that Cfap69 mutant mice showed reduced EOG amplitudes at the highest odorant concentrations, especially when responding to amyl acetate. We have used amyl acetate and heptaldehyde as stimulating odorants for EOG recordings in several previous studies (Song et al., 2008; Cygnar and Zhao, 2009; Stephan et al., 2011; Cygnar et al., 2012; Ferguson and Zhao, 2017) and did not observe any odorant-dependent effect between these two odorants. In previous studies, mutations in OSNs often caused reduced response size and/or slower response kinetics, unlike the Cfap69 mutant. Several things could underlie the observed reduction in the EOG amplitude at high odorant concentrations and the odorant-dependent effect between amyl acetate and heptaldehyde. First, the reduction in the EOG amplitude at high odorant concentrations could reflect a direct effect of the lack of CFAP69 on the olfactory transduction process. Second, it could be due to the integrative nature of the EOG signal. The EOG signal results from the summation of the potential changes of individual responding OSNs in the recording field. The integration of more transient individual signals of the same size could lead to, not only more transient, but also smaller ensembles than the integration of less transient signals, especially when the difference in the width of individual signals become more pronounced. Third, intrinsic odorant receptor properties could account for the odorant-dependent effect because amyl acetate and heptaldehyde should be recognized by different sets of odorant receptors. Future studies, especially at the single-cell level, are needed to delineate the detailed effect of CFAP69 on OSN physiology.
An outstanding question remaining to be answered is what is the mechanism by which CFAP69 regulates olfactory response. Bioinformatic analysis suggests that CFAP69 is an ARM-repeat protein. Proteins containing ARM repeats, including Importin-α and β-catenin, partake in a wide variety of cellular activities and often perform their functions through mediating protein–protein interactions (Groves and Barford, 1999; Hatzfeld, 1999; Andrade et al., 2001). CFAP69 could bind directly or influence indirectly through intermediate(s), one or more of the core transduction components in the olfactory transduction cascade. Therefore, a major effort in future studies should be to identify the interaction partner(s) of CFAP69 to understand how CFAP69 performs its function.
CFAP69 is an evolutionarily conserved protein. We found homologous Cfap69 genes by sequence search in species ranging from unicellular eukaryotes to humans, including all mammalian species searched for and many animals across all major classes of vertebrates. However, we were unable to find the homologous gene in several vertebrate and invertebrate species, including some common model animals such as zebrafish, Drosophila melanogaster, and Caenorhabditis elegans. Because the olfactory transduction process of primary OSNs is conserved throughout the vertebrates, it will be interesting to find how the olfactory system in those vertebrate animals lacking Cfap69 accommodates the lack of a CFAP69-dependent regulatory mechanism.
In addition to the main OE, Cfap69 is also expressed in several other tissues and cells, including the testis and the choroid plexus of the brain (https://www.mousephenotype.org/data/imageComparator?¶meter_stable_id=IMPC_A L Z _0 7 6 _ 001&acc=MGI:2443778). This study represents the first investigation of the function of this novel protein and should provide a reference for continuing studies of CFAP69 function outside of the olfactory system.
Footnotes
This work was supported by the National Institutes of Health (Grant DC007395 to H.Z.) and the Monell Chem Senses Center Fund (Grant G20OD020296 to J.R.). A.K.T. and F.N.D. were partially supported by NIH training grant T32GM007231. We thank Aaron Stephen for the initiation of and the early effort in this study; Randall Reed, Marnie Halpern, Samer Hattar, and Rejji Kuruvilla for critical discussion and suggestions; and members of the Zhao, Hattar, and Kuruvilla laboratories for discussion.
The authors declare no competing financial interests.
References
- Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struct Biol 134:117–131. 10.1006/jsbi.2001.4392 [DOI] [PubMed] [Google Scholar]
- Anholt RR, Aebi U, Snyder SH (1986) A partially purified preparation of isolated chemosensory cilia from the olfactory epithelium of the bullfrog, Rana catesbeiana. J Neurosci 6:1962–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalyar HA, Reed RR (1990) Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science 250:1403–1406. 10.1126/science.2255909 [DOI] [PubMed] [Google Scholar]
- Baumgart S, Jansen F, Bintig W, Kalbe B, Herrmann C, Klumpers F, Köster SD, Scholz P, Rasche S, Dooley R, Metzler-Nolte N, Spehr M, Hatt H, Neuhaus EM (2014) The scaffold protein MUPP1 regulates odorant-mediated signaling in olfactory sensory neurons. J Cell Sci 127:2518–2527. 10.1242/jcs.144220 [DOI] [PubMed] [Google Scholar]
- Belluscio L, Gold GH, Nemes A, Axel R (1998) Mice deficient in G(olf) are anosmic. Neuron 20:69–81. 10.1016/S0896-6273(00)80435-3 [DOI] [PubMed] [Google Scholar]
- Bennett MK, Kulaga HM, Reed RR (2010) Odor-evoked gene regulation and visualization in olfactory receptor neurons. Mol Cell Neurosci 43:353–362. 10.1016/j.mcn.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandawat V, Reisert J, Yau KW (2005) Elementary response of olfactory receptor neurons to odorants. Science 308:1931–1934. 10.1126/science.1109886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billig GM, Pál B, Fidzinski P, Jentsch TJ (2011) Ca2+-activated Cl− currents are dispensable for olfaction. Nat Neurosci 14:763–769. 10.1038/nn.2821 [DOI] [PubMed] [Google Scholar]
- Brunet LJ, Gold GH, Ngai J (1996) General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron 17:681–693. 10.1016/S0896-6273(00)80200-7 [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65:175–187. 10.1016/0092-8674(91)90418-X [DOI] [PubMed] [Google Scholar]
- Buiakova OI, Baker H, Scott JW, Farbman A, Kream R, Grillo M, Franzen L, Richman M, Davis LM, Abbondanzo S, Stewart CL, Margolis FL (1996) Olfactory marker protein (OMP) gene deletion causes altered physiological activity of olfactory sensory neurons. Proc Natl Acad Sci U S A 93:9858–9863. 10.1073/pnas.93.18.9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis RC, Tian H, Wang J, He J, Jiang J, Chen X, Yin W, Connelly T, Ma L, Yu CR, Pluznick JL, Storm DR, Huang L, Zhao K, Ma M (2015) An olfactory cilia pattern in the mammalian nose ensures high sensitivity to odors. Curr Biol 25:2503–2512. 10.1016/j.cub.2015.07.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cygnar KD, Zhao H (2009) Phosphodiesterase 1C is dispensable for rapid response termination of olfactory sensory neurons. Nat Neurosci 12:454–462. 10.1038/nn.2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cygnar KD, Stephan AB, Zhao H (2010) Analyzing responses of mouse olfactory sensory neurons using the air-phase electroolfactogram recording. J Vis Exp 37: pii: 1850. 10.3791/1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cygnar KD, Collins SE, Ferguson CH, Bodkin-Clarke C, Zhao H (2012) Phosphorylation of adenylyl cyclase III at serine1076 does not attenuate olfactory response in mice. J Neurosci 32:14557–14562. 10.1523/JNEUROSCI.0559-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhallan RS, Yau KW, Schrader KA, Reed RR (1990) Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature 347:184–187. 10.1038/347184a0 [DOI] [PubMed] [Google Scholar]
- Dibattista M, Amjad A, Maurya DK, Sagheddu C, Montani G, Tirindelli R, Menini A (2012) Calcium-activated chloride channels in the apical region of mouse vomeronasal sensory neurons. J Gen Physiol 140:3–15. 10.1085/jgp.201210780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G, et al. (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9:e1000582. 10.1371/journal.pbio.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley R, Baumgart S, Rasche S, Hatt H, Neuhaus EM (2009) Olfactory receptor signaling is regulated by the post-synaptic density 95, Drosophila discs large, zona-occludens 1 (PDZ) scaffold multi-PDZ domain protein 1. FEBS J 276:7279–7290. 10.1111/j.1742-4658.2009.07435.x [DOI] [PubMed] [Google Scholar]
- Ferguson CH, Zhao H (2016) Cyclic AMP signaling in the main olfactory epithelium. In: Chemosensory transduction: the detection of odors, tastes, and other chemostimuli (Zufall F, Munger SD, eds), pp 123–140. San Diego: Academic. [Google Scholar]
- Ferguson CH, Zhao H (2017) Simultaneous loss of NCKX4 and CNG channel desensitization impairs olfactory sensitivity. J Neurosci 37:110–119. 10.1523/JNEUROSCI.2527-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein S. (2001) How the olfactory system makes sense of scents. Nature 413:211–218. 10.1038/35093026 [DOI] [PubMed] [Google Scholar]
- Firestein S, Shepherd GM (1991) A kinetic model of the odor response in single olfactory receptor neurons. J Steroid Biochem Mol Biol 39:615–620. 10.1016/0960-0760(91)90259-8 [DOI] [PubMed] [Google Scholar]
- Firestein S, Shepherd GM, Werblin FS (1990) Time course of the membrane current underlying sensory transduction in salamander olfactory receptor neurones. J Physiol 430:135–158. 10.1113/jphysiol.1990.sp018286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein S, Picco C, Menini A (1993) The relation between stimulus and response in olfactory receptor cells of the tiger salamander. J Physiol 468:1–10. 10.1113/jphysiol.1993.sp019756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatpande AS, Reisert J (2011) Olfactory receptor neuron responses coding for rapid odour sampling. J Physiol 589:2261–2273. 10.1113/jphysiol.2010.203687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313:903–919. 10.1006/jmbi.2001.5080 [DOI] [PubMed] [Google Scholar]
- Groves MR, Barford D (1999) Topological characteristics of helical repeat proteins. Current opinion in structural biology 9:383–389. 10.1016/S0959-440X(99)80052-9 [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. (1999) The armadillo family of structural proteins. Int Rev Cytol 186:179–224. [DOI] [PubMed] [Google Scholar]
- Ivic L, Pyrski MM, Margolis JW, Richards LJ, Firestein S, Margolis FL (2000) Adenoviral vector-mediated rescue of the OMP-null phenotype in vivo. Nat Neurosci 3:1113–1120. 10.1038/80632 [DOI] [PubMed] [Google Scholar]
- Jones DT, Reed RR (1989) Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science 244:790–795. 10.1126/science.2499043 [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanageswaran N, Demond M, Nagel M, Schreiner BS, Baumgart S, Scholz P, Altmüller J, Becker C, Doerner JF, Conrad H, Oberland S, Wetzel CH, Neuhaus EM, Hatt H, Gisselmann G (2015) Deep sequencing of the murine olfactory receptor neuron transcriptome. PLoS One 10:e0113170. 10.1371/journal.pone.0113170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Goto T, Sato Y, Katada S, Kinameri E, Yoshihara S, Nishiyori A, Kimura M, Fujita H, Touhara K, Reed RR, Yoshihara Y (2013) Goofy coordinates the acuity of olfactory signaling. J Neurosci 33:12987–12996, 12996a. 10.1523/JNEUROSCI.4948-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB. (2010) Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci 11:188–200. 10.1038/nrn2789 [DOI] [PubMed] [Google Scholar]
- Kerr DS, Von Dannecker LE, Davalos M, Michaloski JS, Malnic B (2008) Ric-8B interacts with G alpha olf and G gamma 13 and co-localizes with G alpha olf, G beta 1 and G gamma 13 in the cilia of olfactory sensory neurons. Mol Cell Neurosci 38:341–348. 10.1016/j.mcn.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Kleene SJ. (2008) The electrochemical basis of odor transduction in vertebrate olfactory cilia. Chem Senses 33:839–859. 10.1093/chemse/bjn048 [DOI] [PubMed] [Google Scholar]
- Kleene SJ, Gesteland RC (1991) Calcium-activated chloride conductance in frog olfactory cilia. J Neurosci 11:3624–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi T, Yau KW (1993) Co-existence of cationic and chloride components in odorant-induced current of vertebrate olfactory receptor cells. Nature 363:71–74. 10.1038/363071a0 [DOI] [PubMed] [Google Scholar]
- Lee AC, He J, Ma M (2011) Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. J Neurosci 31:2974–2982. 10.1523/JNEUROSCI.5067-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ponissery-Saidu S, Yee KK, Wang H, Chen ML, Iguchi N, Zhang G, Jiang P, Reisert J, Huang L (2013) Heterotrimeric G protein subunit Ggamma13 is critical to olfaction. J Neurosci 33:7975–7984. 10.1523/JNEUROSCI.5563-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb BW, Treloar HB, Greer CA (2002) Cell surface carbohydrates reveal heterogeneity in olfactory receptor cell axons in the mouse. Cell Tissue Res 308:7–17. 10.1007/s00441-002-0532-0 [DOI] [PubMed] [Google Scholar]
- Lowe G, Gold GH (1991) The spatial distributions of odorant sensitivity and odorant-induced currents in salamander olfactory receptor cells. J Physiol 442:147–168. 10.1113/jphysiol.1991.sp018787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya DK, Menini A (2014) Developmental expression of the calcium-activated chloride channels TMEM16A and TMEM16B in the mouse olfactory epithelium. Dev Neurobiol 74:657–675. 10.1002/dneu.22159 [DOI] [PubMed] [Google Scholar]
- Mayer U, Küller A, Daiber PC, Neudorf I, Warnken U, Schnölzer M, Frings S, Möhrlen F (2009) The proteome of rat olfactory sensory cilia. Proteomics 9:322–334. 10.1002/pmic.200800149 [DOI] [PubMed] [Google Scholar]
- McClintock TS, Glasser CE, Bose SC, Bergman DA (2008) Tissue expression patterns identify mouse cilia genes. Physiol Genomics 32:198–206. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R (1996) Visualizing an olfactory sensory map. Cell 87:675–686. 10.1016/S0092-8674(00)81387-2 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420:563–573. 10.1038/nature01266 [DOI] [PubMed] [Google Scholar]
- Pietra G, Dibattista M, Menini A, Reisert J, Boccaccio A (2016) The Ca2+-activated Cl- channel TMEM16B regulates action potential firing and axonal targeting in olfactory sensory neurons. J Gen Physiol 148:293–311. 10.1085/jgp.201611622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifferi S, Dibattista M, Menini A (2009) TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflugers Arch 458:1023–1038. 10.1007/s00424-009-0684-9 [DOI] [PubMed] [Google Scholar]
- Ponissery Saidu S, Dibattista M, Matthews HR, Reisert J (2012) Odorant-induced responses recorded from olfactory receptor neurons using the suction pipette technique. J Vis Exp 62:e3862. 10.3791/3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasche S, Toetter B, Adler J, Tschapek A, Doerner JF, Kurtenbach S, Hatt H, Meyer H, Warscheid B, Neuhaus EM (2010) Tmem16b is specifically expressed in the cilia of olfactory sensory neurons. Chem Senses 35:239–245. 10.1093/chemse/bjq007 [DOI] [PubMed] [Google Scholar]
- Reisert J, Matthews HR (1998) Na+-dependent Ca2+ extrusion governs response recovery in frog olfactory receptor cells. J Gen Physiol 112:529–535. 10.1085/jgp.112.5.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert J, Matthews HR (1999) Adaptation of the odour-induced response in frog olfactory receptor cells. J Physiol 519:801–813. 10.1111/j.1469-7793.1999.0801n.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert J, Matthews HR (2001) Response properties of isolated mouse olfactory receptor cells. J Physiol 530:113–122. 10.1111/j.1469-7793.2001.0113m.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert J, Yau KW, Margolis FL (2007) Olfactory marker protein modulates the cAMP kinetics of the odour-induced response in cilia of mouse olfactory receptor neurons. J Physiol 585:731–740. 10.1113/jphysiol.2007.142471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AJ, et al. (2005) Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet 37:1135–1140. 10.1038/ng1644 [DOI] [PubMed] [Google Scholar]
- Sammeta N, Yu TT, Bose SC, McClintock TS (2007) Mouse olfactory sensory neurons express 10,000 genes. J Comp Neurol 502:1138–1156. 10.1002/cne.21365 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Scott-Johnson PE (2002) The electroolfactogram: a review of its history and uses. Microsc Res Tech 58:152–160. 10.1002/jemt.10133 [DOI] [PubMed] [Google Scholar]
- Sinnarajah S, Dessauer CW, Srikumar D, Chen J, Yuen J, Yilma S, Dennis JC, Morrison EE, Vodyanoy V, Kehrl JH (2001) RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature 409:1051–1055. 10.1038/35059104 [DOI] [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. San Francisco: W.H. Freeman. [Google Scholar]
- Song Y, Cygnar KD, Sagdullaev B, Valley M, Hirsh S, Stephan A, Reisert J, Zhao H (2008) Olfactory CNG channel desensitization by Ca2+/CaM via the B1b subunit affects response termination but not sensitivity to recurring stimulation. Neuron 58:374–386. 10.1016/j.neuron.2008.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AB, Shum EY, Hirsh S, Cygnar KD, Reisert J, Zhao H (2009) ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc Natl Acad Sci U S A 106:11776–11781. 10.1073/pnas.0903304106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AB, Tobochnik S, Dibattista M, Wall CM, Reisert J, Zhao H (2011) The Na(+)/Ca(2+) exchanger NCKX4 governs termination and adaptation of the mammalian olfactory response. Nat Neurosci 15:131–137. 10.1038/nn.2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 101:6062–6067. 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadenev AL, Kulaga HM, May-Simera HL, Kelley MW, Katsanis N, Reed RR (2011) Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc Natl Acad Sci U S A 108:10320–10325. 10.1073/pnas.1016531108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotier D. (1994) Intensity coding in olfactory receptor cells. Semin Cell Biol 5:47–54. 10.1006/scel.1994.1007 [DOI] [PubMed] [Google Scholar]
- Trotier D, MacLeod P (1983) Intracellular recordings from salamander olfactory receptor cells. Brain Res 268:225–237. 10.1016/0006-8993(83)90488-2 [DOI] [PubMed] [Google Scholar]
- Von Dannecker LE, Mercadante AF, Malnic B (2005) Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Galphaolf. J Neurosci 25:3793–3800. 10.1523/JNEUROSCI.4595-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, Xia Z, Gold GH, Storm DR (2000) Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron 27:487–497. 10.1016/S0896-6273(00)00060-X [DOI] [PubMed] [Google Scholar]
- Yan C, Zhao AZ, Bentley JK, Loughney K, Ferguson K, Beavo JA (1995) Molecular cloning and characterization of a calmodulin-dependent phosphodiesterase enriched in olfactory sensory neurons. Proc Natl Acad Sci U S A 92:9677–9681. 10.1073/pnas.92.21.9677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou DJ, Chesler AT, Le Pichon CE, Kuznetsov A, Pei X, Hwang EL, Firestein S (2007) Absence of adenylyl cyclase 3 perturbs peripheral olfactory projections in mice. J Neurosci 27:6675–6683. 10.1523/JNEUROSCI.0699-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Molecules as documents of evolutionary history. J Theor Biol 8:357–366. 10.1016/0022-5193(65)90083-4 [DOI] [PubMed] [Google Scholar]