FIGURE 6. Mouse IFN-β helix 4 induces membrane curvature in bacterial membranes and kills S. aureus.
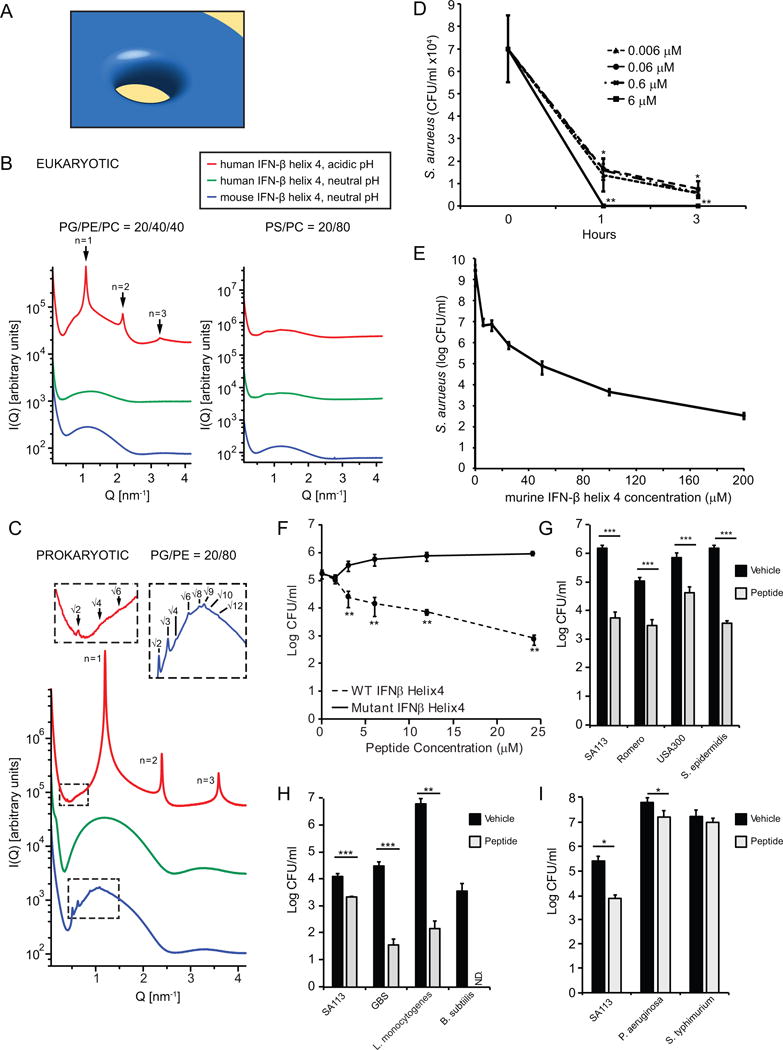
(A) Model of negative Gaussian curvature (NGC) required for membrane destabilization processes such as pore formation by AMPs. (B) IFN-β helix 4 does not generate NGC in eukaryotic-like membranes. SAXS spectra for mouse and human IFN-β helix 4 with eukaryotic model membranes DOPG/DOPE/DOPC = 20/40/40 and DOPS/DOPC = 20/80 at P/L = 1/50 peptide to lipid (P/L) molar ratio. (C) IFN-β helix 4 generates NGC in prokaryotic-like membranes. SAXS spectra from prokaryotic model membrane, DOPG/DOPE = 20/80, vesicles incubated with either mouse or human IFN-β helix 4 at P/L = 1/50 molar ratio. Insets show expanded view of cubic phase reflections. (D) S. aureus were incubated with VEH or mIFN-β helix 4 at 0.006, 0.06, 0.6, and 6 μM. Bacteria and treatments were incubated for 1 or 3 hours. Remaining bacteria were serially-diluted and plated for enumeration. (E) S. aureus were incubated with VEH or mouse IFN-β helix 4 at 6.25, 12.5, 25, 50, 100, and 200 μM. Bacteria and treatments were incubated for 3 hours. Remaining bacteria were serially-diluted and plated for enumeration. (F) Wild-type (WT) and mutant IFN-β helix 4 peptide were incubated with 1×105 SA113/ml in buffer + 1%THB for 3 hours. Remaining bacteria were serially-diluted and plated for enumeration. (G–I) Bacteria (1×105/ml) were incubated with vehicle or 6 μM mouse hexlix 4 peptide for 3 hours at 37°C. Remaining bacteria were serially-diluted and plated for enumeration. Data are shown as mean ± SD, * p ≤ 0.05,** p ≤ 0.01, ***p≤0.001 (unpaired, two-tailed t test). N.D.- below limit of detection.
