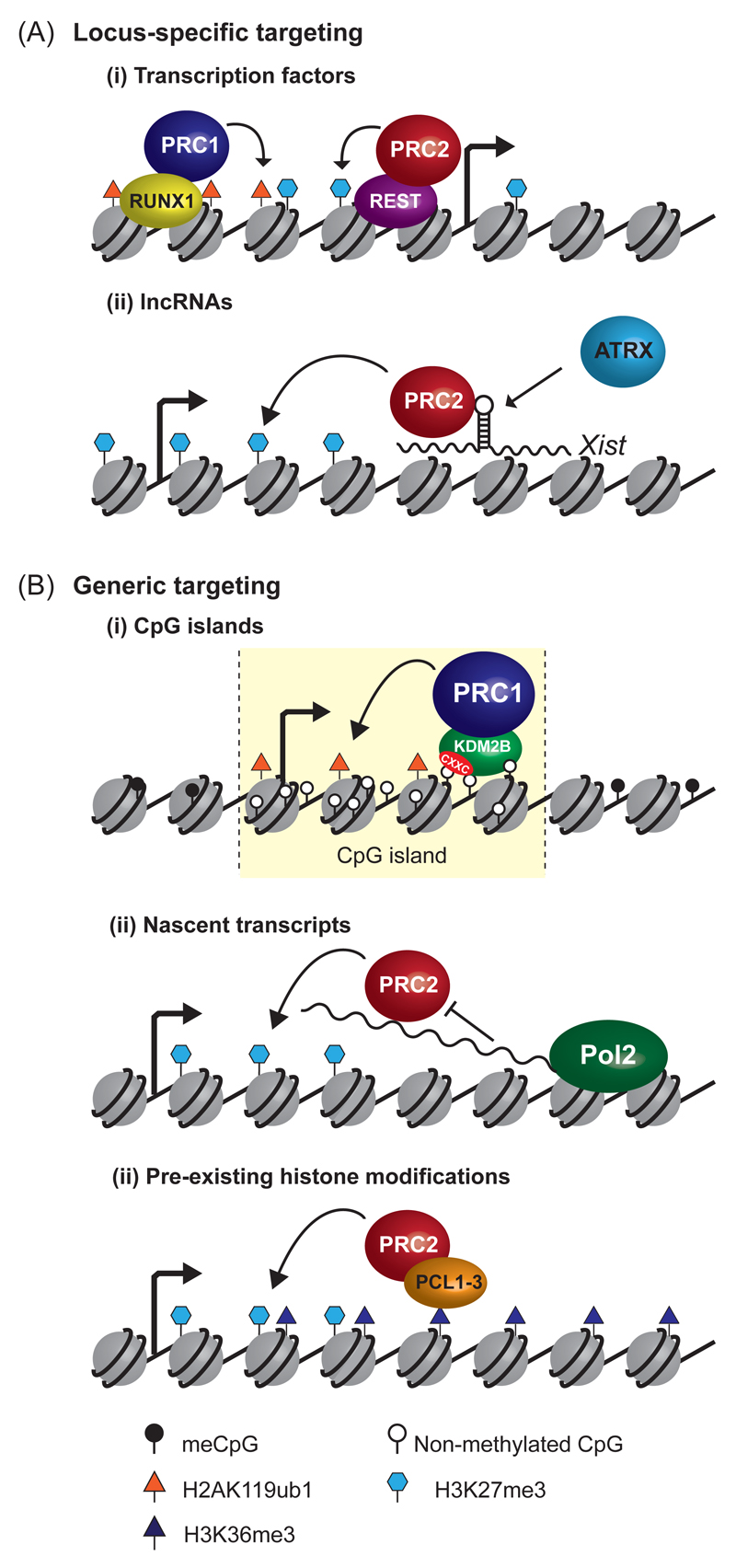
Figure 1. Getting polycomb repressive complexes to chromatin.
a | Locus-specific targeting of polycomb repressive complexes. PRC1 and PRC2 can associate with DNA binding transcription factors (top-left), such as RUNX1 and REST, which guide these complexes to chromatin. Similarly, interactions with long non-coding RNAs such as Xist function in chromosome- and locus-specific targeting of polycomb complexes (bottom-left). The chromatin remodelling protein ATRX may remodel the structure of Xist to achieve interaction with PRC2. Following recruitment to chromatin, PRC1 catalyses ubiquitylation of H2AK119 and PRC2 catalyses trimethylation of H3K27, as indicated by rounded arrows. The square arrows indicate transcription start sites.
b | Generic targeting of polycomb repressive complexes to gene regulatory elements. A variant PRC1 complex containsthe KDM2B protein. KDM2B has a zinc-finger CxxC DNA binding domain (red area) that specifically recognizes non-methylated CpG dinucleotides. This allows KDM2B to bind at CpG islands genome-wide, and contributes to PRC1 occupancy at these elements (top-right). PRC2 interacts with nascent RNA polymerase II (Pol II) transcripts at 5’-ends of genes, which may provide a mechanism to maintain repression at silent genes following stochastic transcription initiation events. Alternatively, at active genes, interaction of PRC2 with nascent RNA may constrain the catalytic activity of PRC2 and protect against polycomb-mediated repression (middle-right). A subset of PRC2 complexes contain PCL proteins which bind to H3K36me3, a modification associated with active transcription. This may enable PRC2 to bind at, or spread into, previously transcribed regions, catalysing H3K27me3 at these regions.