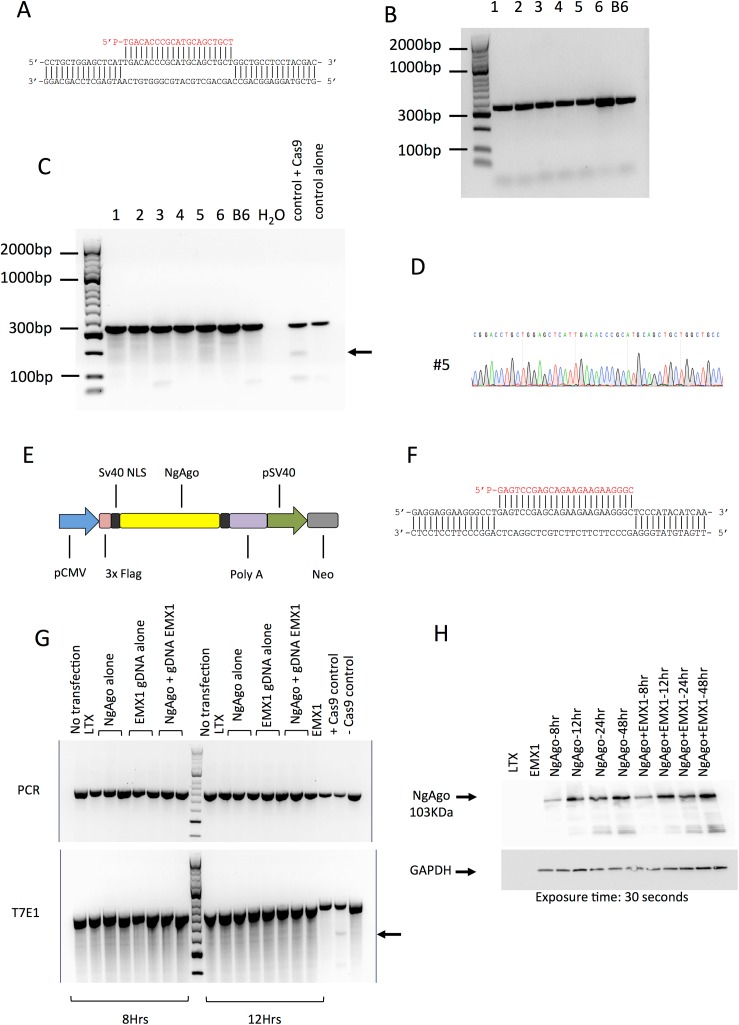
Fig 1. No evidence for double strand break cleavage and editing from NgAgo.
(A) DNA sequence indicating the locus targeted for the exon 26 of Sptb. The gDNA sequence is indicated in red. (B) Gel electrophoresis (1.5%) of Sptb blastocysts (n = 6) co-injected with nls-NgAgo-GK plasmid (2.5 ng/μl) and 2.5 ng/μl of gDNA. C57BL/6 DNA (B6) was also amplified as a control. The PCR product is 326 bp. (C) T7 endonuclease 1 (T7E1) assay on the Sptb blastocysts indicating the absence of heteroduplexes suggesting indels in 6 blastocysts co-injected with nls-NgAgo-GK and gDNA. C57BL/6 (B6) non-edited control DNA was utilized as a negative control. A positive control in mouse zygotes edited with CRISPR/Cas9 was used as a positive control. The arrows indicate the presence of heteroduplexes suggesting a successful editing of the DNA. (D) Representative chromatogram of one Sptb blastocyst (#5) suggesting no editing of the DNA under the DNA-guided NgAgo. (E) Schematic diagram representing the flag-nls-NgAgo-GK plasmid. The expression of NgAgo is driven from a CMV promoter. A Flag tag was inserted in the 5’ end of NgAgo sequence. Two Sv40 nuclear localization signals were inserted in the 3’ end of the NgAgo sequence and in the 3’ end of the flag tag. A Ploy A tail was appended to the sequence in the 3’ end a Neomycin cassette was added in the 3’ end of the plasmid sequence. (F) DNA sequence indicating the targeting of the exon 3 from EMX1 human sequence. The gDNA sequence is indicated in red. (G) Gel electrophoresis (2%) of the PCR for EMX1 in HEK293T cells at 8 and 12 hours post lipofection. The control samples were: The DNA without transfection, the lipofection reagent (LTX) and EMX1 DNA. The HEK293T cells were transfected with NgAgo alone, EMX1 gDNA alone or co-transfected with NgAgo and EMX1 gDNA. A control DNA was successfully edited with CRISPR/Cas9 (+ Cas9 control) and was utilized as a negative control (- Cas9 control). The top electrophoresis gel respresents the PCR only whereas the bottom gel represents the T7E1 assay. The arrows indicate the formation of heteroduplexes for CRISPR/Cas9 genome editing. (H) Western blot of NgAgo protein production with and without the co-transfection of the gDNA from 8 to 48 hours post lipofection. The staining was performed with a monoclonal Flag anti-antibody and anti-GAPDH anti-antibody. The top band represents NgAgo at 103KDa. GAPDH was utilized as a Housekeeper gene. The exposure time was 30 seconds. The controls were the lipefection agent alone (LTX), lane 1 and the gDNA EMX1 alone (lane 2). The smears under the NgAgo band show the degradation of the protein stained with the Flag tag.