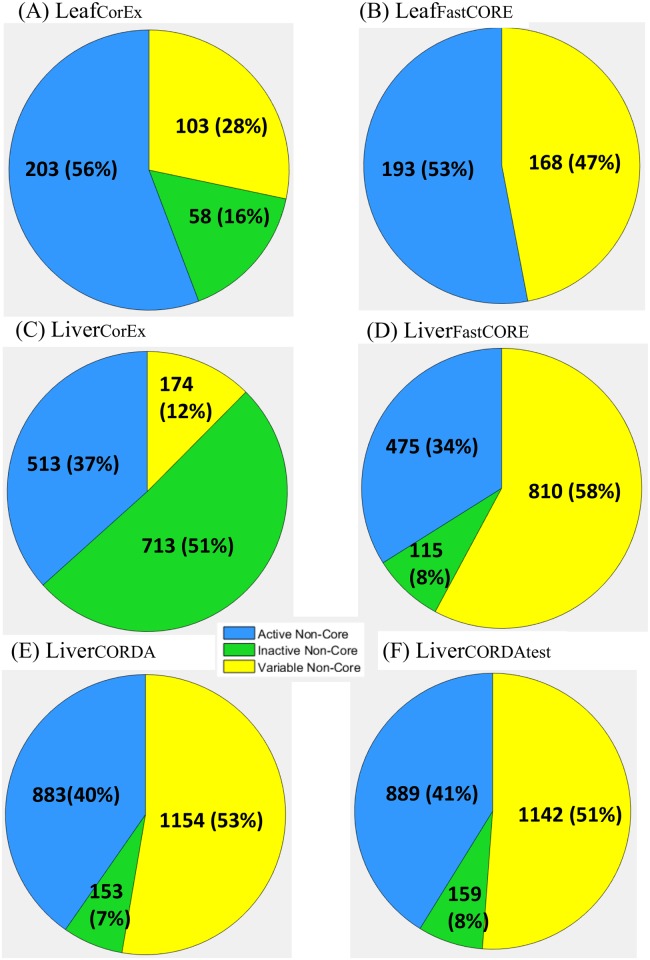
Fig 4. Alternative optima of CorEx and FastCORE context-specific network extractions.
The results are divided into the leaf-specific scenario for the CorEx (A) and FastCORE (B) alternative optima, and the liver-specific scenario, for CorEx (C), FastCORE (D) and CORDA without applying the metabolic test (E) and applying the metabolic test (F) to further constraint the alternative optima space (see main text). In all cases, non-core reactions are partitioned into the set that is always included in all alternative networks, (the fixed non-core set, in green), the set that is always excluded (excluded non-core, grey) and the variable non-core set (yellow) which is formed by reactions that are included in some of the alternative networks. In both, the leaf- and the liver-specific scenario, the alternative optima networks generated by CorEx contain a larger proportion of fixed non-core reactions and a smaller proportion of variable non-core reactions. These differences in behavior may be explained by the greater number of non-core reactions that are added by FastCORE, as compare to CorEx, in the optimal solution (see main text).