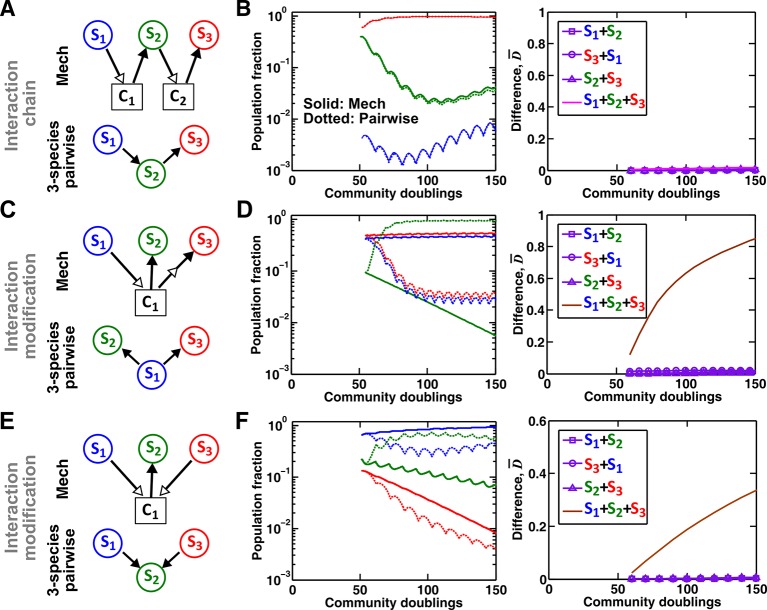
Figure 7. Interaction chain but not interaction modification may be represented by a multispecies pairwise model.
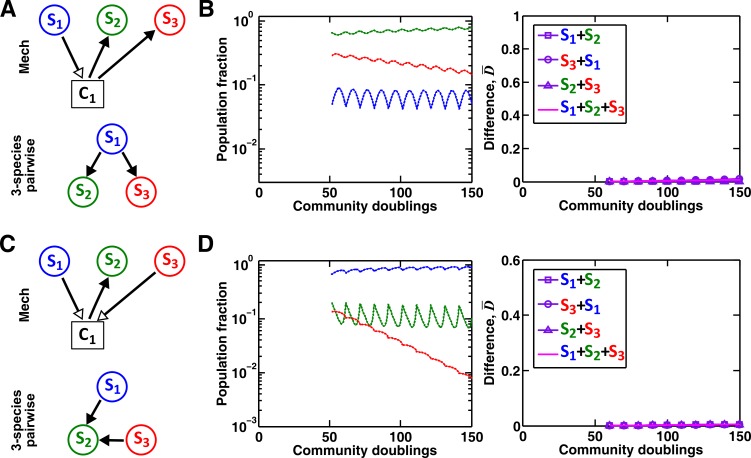
We examine three-species communities engaging in indirect interactions. Each species pair is representable by a two-species pairwise model (saturable L-V or alternative pairwise model, purple in the right columns of B, D, and F). We then use these two-species pairwise models to construct a three-species pairwise model, and test how well it predicts the dynamics known from the mechanistic model. In B, D, and F, left panels show dynamics from the mechanistic models (solid lines) and three-species pairwise models (dotted lines). Right panels show the difference metric . (A–B) Interaction chain: S1 affects S2, and S2 affects S3. The two interactions employ independent mediators C1 and C2, and both interactions can be represented by the saturable L-V pairwise model. The three-species pairwise model matches the mechanistic model in this case. Simulation parameters are provided in Figure 7—source data 1. (C–F) Interaction modification. In both cases, the three-species pairwise model fails to predict reference dynamics even though the dynamics of each species pair can be represented by a pairwise model. (C–D) S3 consumes C1, a mediator by which S1 stimulates S2. Parameters are listed in Figure 7—source data 2. Here, S1 changes the nature of interaction between S2 and S3: S2 and S3 do not interact in the absence of S1, but S3 inhibits S2 in the presence of S1. The three-species pairwise model makes qualitatively wrong prediction about species coexistence. As expected, if S3 does not remove C1, the three-species pairwise model works (Figure 7—figure supplement 1A–B). (E–F) S1 and S3 both supply C1 which stimulates S2. Here, no species changes ‘the nature of interactions’ between any other two species: both S1 and S3 contribute reusable C1 to stimulate S2. S1 promotes S2 regardless of S3; S3 promotes S2 regardless of S1; S1 and S3 do not interact regardless of S2. However, a multispecies pairwise model assumes that the fitness effects from the two producers on S2 will be additive, whereas in reality, the fitness effect on S2 saturates at high . As a result, the three-species pairwise model qualitatively fails to capture relative species abundance. As expected, if C1 affects S2 in a linear fashion, the community dynamics is accurately captured in the multispecies pairwise model (Figure 7—figure supplement 1C–D). Simulation parameters are listed in Figure 7—source data 3.
DOI: http://dx.doi.org/10.7554/eLife.25051.026