Abstract
Rapid and affordable tumor profiling has led to an explosion of genomic data that is facilitating the development of new cancer therapies. The potential of therapeutic strategies aimed at inactivating the oncogenic lesions that contribute to the aberrant survival and proliferation of tumor cells has yielded remarkable success in some malignancies such as BRAF-mutant melanoma and BCR-ABL expressing chronic myeloid leukemia. However, the direct inhibition of several well-established oncoproteins in some of these cancers is not possible or produces only transient benefits. Functional genomics represents a powerful approach for the identification of vulnerabilities linked to specific genetic alterations and has provided substantial insights into cancer signaling networks. Still, as inhibition of gene function can have diverse effects on both tumor and normal tissues, information on the potency of target inhibition on tumor growth as well as the toxic side effects of target inhibition are also needed. Here, we discuss our RNA interference (RNAi) pipeline for cancer target discovery based on our optimized short-hairpin RNA (shRNA) tools for negative selection screens and inducible RNAi platform that, in combination with embryonic stem cell (ESC)-based genetically engineered mouse models (GEMMs), enable deep in vivo target validation.
TARGET IDENTIFICATION AND VALIDATION IN ONCOLOGY
Cancer genomes have complex landscapes of mutations and diverse types of genetic aberrations (Lawrence et al. 2013). Advances in sequencing technologies have greatly facilitated genome characterization, making it possible to compile a nearly complete catalog of genes altered in human tumors and the development of new therapies that rationally exploit these alterations. In a number of cases where a cancer driver has been functionally validated, particularly in the case of certain activated oncogenes, therapeutic strategies to inhibit the oncogenic event have proven remarkably effective in treating patients with these lesions. However, not all oncogenic events are druggable by traditional pharmacologic approaches, and this general approach cannot exploit loss of tumor suppressors. As a consequence, genome characterization alone has not identified obvious targets for a large fraction of human cancers. Therefore, it seems likely that the combination of genomic and functional approaches will be needed to identify new cancer vulnerabilities that lead to the rational development of effective therapies.
In principle, an ideal cancer target would be required for the ongoing proliferation and/or survival of cancer cells but not normal tissues. Such targets can be classified into three general categories: (1) the cancer “driver” itself, whose direct inhibition can produce an anticancer effect; (2) a “client” of a cancer driver that mediates its tumor-promoting effect and whose inhibition achieves a similar outcome as would targeting the driver itself; and (3) a “synthetic lethal” target that reflects an induced dependency of a cancer cell harboring a particular driving genetic event. Functional genomic approaches are ideal for identifying targets in classes 2 and 3. Still, even when a genotype-specific vulnerability is identified, it remains to be determined how robust target inhibition will be in an in vivo context and whether there may be deleterious effects to normal tissues. As discussed below, functional genomic techniques and new animal modeling approaches provide one path forward.
RNA interference (RNAi) technology is a powerful tool for identifying genes whose suppression affects cell proliferation and viability. Although genome-wide RNAi screens have been widely used for the identification of new therapeutic targets in cancer, these screens are technically challenging, tedious, and error-prone because of the high complexity of the short-hairpin RNA (shRNA) libraries. During the past few years, we have incorporated an RNAi platform optimized for negative selection screens that consists of three modular components. First, we use miR30-based shRNA systems that enable robust gene knockdown from constitutive and inducible promoters and that link the shRNAs to fluorescent reporters, allowing the facile tracking and/or sorting of shRNA-expressing cells (Hemann et al. 2003; Dickins et al. 2005, 2007; Zuber et al. 2011a). This tracking system significantly increases the sensitivity of our screens by reducing the noise derived from cells that do not efficiently induce the shRNA. Second, we take a systematic approach to identify shRNAs optimized for potency and specificity (Fellmann et al. 2011). Third, we use focused shRNA libraries targeting genes hypothesized to be important for the desired phenotype, which reduces the library complexity and increases the likelihood of obtaining robust and reproducible results.
Using this conceptual approach, we performed several in vivo RNAi screens and identified a series of relevant tumor suppressors (Zender et al. 2008; Bric et al. 2009; Scuoppo et al. 2012). We also identified Brd4 as crucial for the maintenance of acute myeloid leukemia (AML) and platelet-derived growth factor receptor beta (PDGFRβ) as an antimetastasis target for tumors harboring mutant p53 (Zuber et al. 2011b; Weissmueller et al. 2014). More recently, we uncovered a synthetic lethal interaction between MYC-overexpressing hepatocellular carcinomas (HCCs) and CDK9 inhibition, and identified that fibroblast growth factor receptor 1 (FGFR1) inhibition confers hypersensitivity to MEK inhibition in KRAS-mutant lung and pancreatic cancer cells (Huang et al. 2014; Manchado et al. 2016). Importantly, all of the principles used in these studies can be directly applied to functional genomic approaches using variations on clustered regularly interspaced short palindromic repeat (CRISPR)–Cas9 genome editing technology.
A key aspect in the process of identifying a new cancer target is validating whether the target is required for the maintenance of the cancer phenotype in the complex physiological setting of in vivo tumor development. Mouse models represent one “gold standard” system for validating candidate targets. Tumor cell line xenograft models have been widely used for in vivo validation of candidate targets, because of their tractability, flexibility, and potential to reflect the genetic diversity of human tumors. However, the recipient mice in these subcutaneous graft models are immune-deficient and the tumor grows in an atypical site, thus not capturing the complex interaction between tumor cells, stroma, and immune system. To partially address these issues, our pipeline incorporates an approach to rapidly transit hits from a genetic screen to in depth in vivo target validation using genetically and pathologically accurate cancer mouse models.
Our validation approach is based on our inducible shRNA platform that enables suppressing gene function in established tumors instead of at the time of cell engraftment or, in genetically engineered models, at tumor initiation. The most elegant iteration of the method is based on the derivation of ESC lines that harbor multiple conditional cancer-predisposing alleles, tissue-specific Cre recombinases, and reverse tet-transactivators that, in combination, are used directly to generate a functional cancer mouse model without the extensive strain intercrossing required with conventional methods. These models allow us to not only genetically validate candidate targets in a physiologic context and with an unprecedented depth but also to anticipate potential toxicities associated with the therapeutic strategy, because the target is inhibited in the majority of tissue types and not just in the tumor cells.
Here, we summarize our RNAi pipeline that uses, in many cases, the same reagents to seamlessly go from genetic screens for target identification to deep in vivo validation approaches that stand in contrast to previous efforts that have simply looked at genetic dependencies in cell lines or xenograft models. We first discuss and illustrate our RNAi screening approach for target discovery by describing two shRNA screens aimed at identifying vulnerabilities in Myc-driven HCC and mechanisms of resistance to MEK inhibition in KRAS-mutant lung cancer. We then summarize our GEMM-ESC platform for in depth in vivo target validation and for anticipating toxicities associated to target inhibition.
RNAi SCREENS FOR TARGET IDENTIFICATION
Much of the current thinking in the identification and development of novel cancer targets involves the concept of precision medicine—specifically the concept of matching patients’ tumors with specific genetic configurations and drug treatments predicted to target those lesions. The potential of such strategies has been revealed by the success of imatinib for BCR-ABL-expressing chronic myeloid leukemia and gefitinib or erlotinib for epidermal growth factor receptor (EGFR)-mutant lung adenocarcinoma. However, for some genetic lesions (e.g., amplified MYC or mutant KRAS), this has not been possible.
Functional genomics provides a nonbiased approach to uncover vulnerabilities linked to specific genetic alterations and has been widely applied toward this goal over the last decade. In its most basic iteration, RNAi or CRISPR libraries are introduced into cells and the representation of individual shRNAs or single-guide RNAs (sgRNAs) is assessed by next-generation sequencing, with the expectation that cells expressing shRNAs or sgRNAs targeting genes required for proliferation or survival will be depleted with time or under specific conditions (e.g., drug treatments).
Like many laboratories, we use functional genomics to identify cancer drug targets, focusing on exploiting vulnerabilities produced by important but undruggable cancer drivers. Below, we summarize our efforts to optimize shRNA screens for the identification of cancer therapeutic targets and illustrate this approach with two recent screens performed in our laboratory, which identified (i) CDK9—a key component of the positive transcription elongation factor b (P-TEFb) complex—as required for the proliferation of MYC-overexpressing HCC (Huang et al. 2014) and (ii) FGFR1 as a key mediator of a feedback signaling loop that induces adaptive resistance to MEK inhibition in KRAS-mutant lung cancer cells (Manchado et al. 2016).
Our screening approach strives to reduce library complexity and efficiently produces customized shRNA libraries in vectors appropriate for the screen. Using low complexity libraries makes screenings technically easier and more cost effective by requiring less cell culture, and increases the likelihood of identifying significant targets by allowing more replicates and enabling more accurate shRNA quantitation than when performing error-prone genome-wide screens. To achieve this, we generate focused lists of gene candidates based on the biology of the system that we would like to interrogate. For example, to identify new therapeutic strategies in MYC-overexpressing HCCs, we generated a customized shRNA library capable of suppressing proteins for which small molecule inhibitors are available (Huang et al. 2014). For identifying sensitizers of KRAS-mutant lung cancer cells to MEK inhibition, we screened a kinase library by hypothesizing that the activation of kinase signaling cascades mediates signaling reactivation and adaptive resistance to MEK inhibitors (Manchado et al. 2016). By taking this focused approach, we were able to follow up positive hits with relevant functional experiments, such as using small molecule inhibitors predicted to mimic genetic perturbations.
In addition to developing a biological hypothesis to focus our screen, we have invested heavily in optimizing tools and methods to produce robust results. Indeed, a multiplexed RNAi screen requires shRNAs that efficiently suppress their target and a system that allows the efficient delivery and expression of the shRNAs in the target cells. To meet this requirement, we developed an algorithm derived from analyzing data from a biological reporter assay that functionally evaluates shRNA potency on a large scale (Fellmann et al. 2011). To minimize false negatives, enhance the ability to study the loss of function of proliferation and survival genes, and provide a feasible system to efficiently deliver the shRNAs, we developed optimized systems such as the TRMPV tet-responsive miR30-based retroviral vector (Zuber et al. 2011a). This vector enables inducible shRNA expression and, by coupling the shRNA to a fluorescent reporter, allows only those cells expressing the shRNA to be analyzed, increasing the sensitivity of our screens. By using this approach, we produced kinase and drug target gene shRNA libraries that harbor remarkably accurate shRNA sequences of relatively equal distribution, which increases the likelihood of obtaining robust results (Huang et al. 2014; Manchado et al. 2016).
To identify targets specifically required for the proliferation/ survival of Myc-overexpressing murine HCC cells or synthetic lethal interactions with the MEK inhibitor trametinib in human KRAS-mutant lung cancer cells, the vector libraries described above (human kinome and murine “drug target” genes) containing inducible shRNAs and a neomycin resistance gene were transduced in triplicate into either Myc-overexpressing or KRAS-mutant cancer cells under conditions that predominantly result in a single retroviral integration per cell. Our shRNA libraries included six informatically predicted shRNAs per gene and shRNAs targeting “neutral” and “essential” genes (Huang et al. 2014; Manchado et al. 2016), which provide negative and positive controls, respectively, help to calibrate sequencing results and normalize between independent experiments. Upon shRNA transduction and selection with neomycin, cells were treated with doxycycline (to allow shRNA induction) or with doxycycline together with trametinib to identify synthetic lethal interactions with MEK inhibition in KRAS-mutant cells, and were cultured for 10 population doublings under conditions such that each shRNA was represented in at least 1000 cells through the course of the experiment.
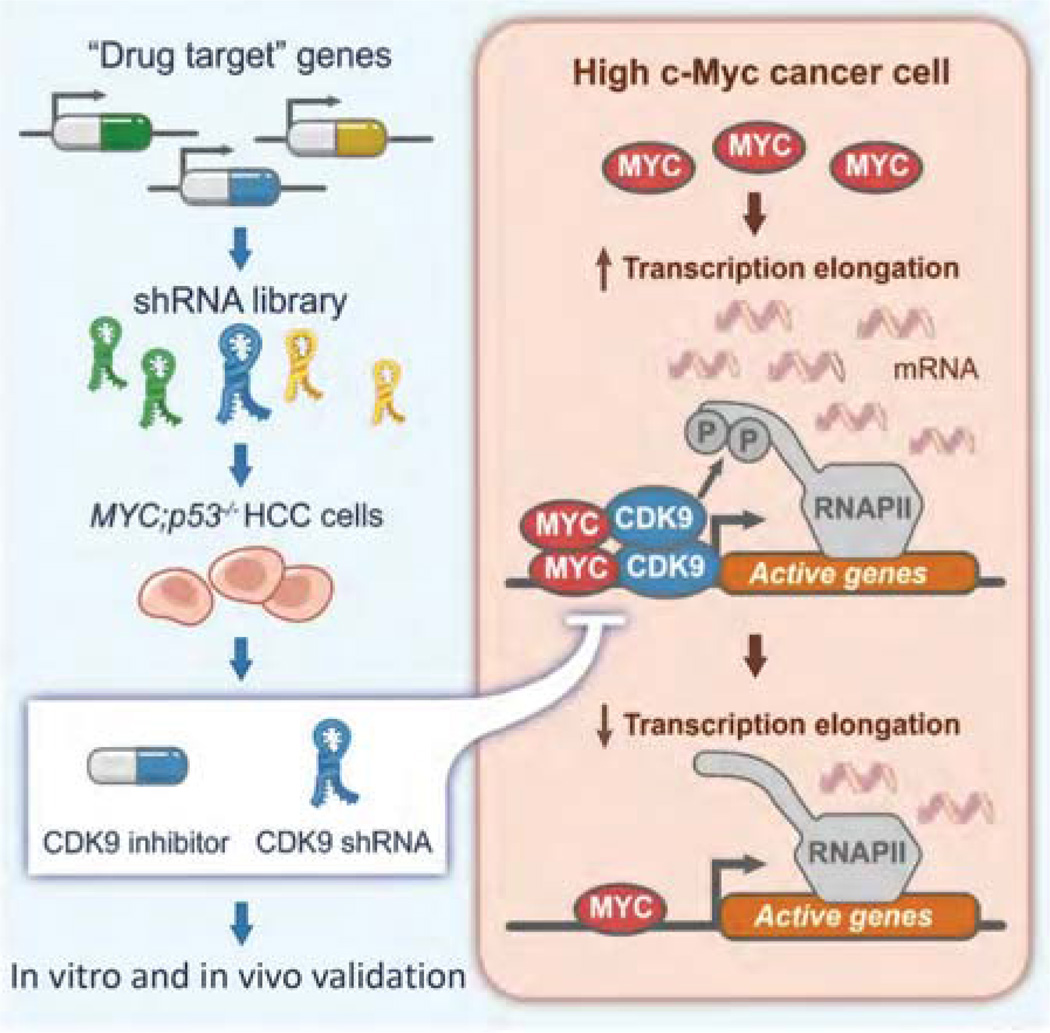
To determine shRNA representation, shRNA cassettes were amplified from genomic DNA and quantified by deep sequencing. Importantly, the use of a vector that links the shRNA to a fluorescent reporter allowed us to evaluate only those cells in which the shRNA was expressed (by fluorescence-activated cell sorting) and eliminated sequencing noise from silenced elements. Upon sequencing, data were analyzed using a custom script and the fold change of shRNA representation was calculated by comparing normalized shRNA reads at tf to normalized reads at t0, after eliminating from the analysis those shRNAs underrepresented in the initial screen samples. As a general rule, we require that candidate genes for further validation have at least two shRNAs that show more than fourfold average depletion and FDR < 0.05. Following this precise method we identified Cdk9, Psmb2, and Mcm6 as potential targets in Myc-overexpressing HCC and FGFR1 and other mitogen-activated protein kinase (MAPK) signaling effectors as sensitizers of KRAS-mutant lung cancer cells to MEK inhibition (Fig. 1; Huang et al. 2014; Manchado et al. 2016).
Figure 1.
Schematic of a focused RNAi screen for genes encoding known drug targets. By screening a custom library of short-hairpin RNAs (shRNAs) directed toward known drug targets in a genetically defined Myc-driven hepatocellular carcinoma (HCC) model, we identified CDK9—a key component of the positive transcription elongation factor b (P-TEFb) complex—as required for MYC-overexpressing HCC. Results from shRNA-mediated and pharmacological CDK9 inhibition together highlight the relevance of transcription elongation in the addiction of cancer cells to MYC and suggest that targeting MYC-mediated transcription elongation can be an effective strategy for the treatment of MYC-overexpressing tumors.
On the basis of the above experiences, we believe that our focused shRNA libraries and screening pipeline are capable of generating robust and reproducible results in both murine and human systems. By screening shRNA libraries targeting known drug target genes and human kinases in a murine HCC model driven by Myc overexpression and p53 loss and in a KRAS-mutant lung cancer cell line, respectively, we described a critical role for CDK9 in MYC-dependent transcription elongation and HCC maintenance and showed that the MEK inhibitor trametinib provokes a compensatory response involving FGFR1 that leads to adaptive drug resistance (Huang et al. 2014; Manchado et al. 2016).
It is noteworthy that the emergence of CRISPR–Cas9 technologies (Doudna and Charpentier 2014) has greatly facilitated functional genomics efforts (Shalem et al. 2015). Importantly, it is becoming increasingly clear that CRISPR–Cas9 technologies represent a powerful orthogonal approach to the RNAi methods described above (Deans et al. 2016). This method, in its simplest form, involves the CRISPR-associated Cas9 endonuclease and a sgRNA, can be experimentally harnessed to target and cleave DNA in an exquisite sequence-specific manner (Doudna and Charpentier 2014). One major advantage of CRISPR is that a much larger fraction of sgRNAs is effective when compared with shRNAs, making it less likely to have false-negative results (Morgens et al. 2016). Also, unlike RNAi, CRISPR–Cas can produce true null alleles. Additional iterations of the CRISPR– Cas methodology using nuclease-deficient Cas9 can enable inducible and reversible gene activation and repression (Shalem et al. 2015). Although these methods are still in their infancy, they will undoubtedly further enhance the capabilities of functional genomics screens.
Still, for many therapeutic targets, hypomorphic effects from RNAi are desirable and can help identify a realistic therapeutic window for inhibition of essential gene products. Recent efforts to increase the knockdown efficiency of miR-30-based shRNAs led to an enhanced microRNA-based hairpin, termed miR-E (Fellmann et al. 2013). The utility of single-copy miR-E shRNAs to facilitate the functional annotation of the genome has also been evaluated and validated in multiple independent studies (Huang et al. 2014; Schwickert et al. 2014; Watanabe et al. 2016). Moreover, new algorithms for microRNA-based shRNA technologies are also increasing the fraction of functional shRNAs in libraries (Pelossof et al., unpubl.). Consequently, our and other laboratories (Deans et al. 2016) are currently using RNAi and CRISPR as two orthogonal approaches that reinforce each other and believe that such an integrative approach will maximize the information obtainable from functional genomics screens.
RNAi FOR TARGET VALIDATION: IN VITRO AND IN VIVO APPROACHES
In Vitro Target Validation
A critical step in the process of target discovery involves the validation of the potential cancer targets. In our screening pipeline, target candidates identified following our scoring criteria are validated through several methods. First of all, candidate shRNAs are individually tested in competitive in vitro proliferation assays together with positive control shRNAs targeting genes such as PLK1, RPA1, and the oncogenic drivers (i.e., Myc and KRAS), and negative control shRNAs targeting Renilla luciferase. To partially recapitulate the settings of the screen, shRNA-transduced cells expressing a fluorescent reporter are mixed with nontransduced cells in an equal proportion and the percentage of each population is quantified over time by flow cytometry.
Next, to assess the absolute effect of target inhibition on cell proliferation and survival, we test the effect of candidate shRNAs in clonogenic and apoptosis assays in multiple cell lines. For shRNAs identified as synthetic lethal partners with a drug, we also examine whether the effects are additive or synergistic using different approaches such as the Chou–Talalay or Loewe methods (Loewe 1953; Chou 2010). For situations where small molecule inhibitors targeting the candidate gene exist, we test whether the drug recapitulates the effect of the shRNAs. We examine how the drug affects the proliferation and survival of cancer cells by using in vitro clonogenic assays and by measuring apoptosis (annexin V/propidium iodide), and we examine the synergistic activity of the drug combination using the methods mentioned above. Highlighting the robustness of our screening approach, we usually validate ~80% of the candidate genes identified following the scoring criteria described above.
As we aim to identify targets whose inhibition show selective antiproliferative effects in cancer cells or in cells from a certain tumor type, we examine the effect of suppressing the positive hits in nontransformed cells and in cancer cells from different lineages or with different genetic drivers. For example, we validated the antiproliferative effects of CDK9 inhibition in Myc-overexpressing HCC cells, but observed a limited effect in nontransformed liver cells (Huang et al. 2014). Similarly, FGFR1 inhibition showed combined activity with trametinib in KRAS-mutant pancreatic and lung cancer cells, but not in KRAS wild-type lung cancer cells or KRAS-mutant colorectal lines (Manchado et al. 2016).
An important aspect of target validation is confirming that the phenotypes induced by shRNAs are due to ontarget effects. To do this, we use several orthogonal methods: (i) We validate our results in multiple cancer cell lines harboring different genetic drivers and derived from different tumor tissue types; (ii) we ensure that multiple shRNAs capable of knocking down the target protein produce similar results; (iii) we confirm that CRISPR–Cas9 genome editing targeting of the candidates recapitulates the shRNA effects; (iv) if available, we test and compare the effect of small molecule inhibitors targeting the hit; and (v) we perform rescue experiments by expressing the cDNA of the candidate gene containing a silent mutation in the shRNA target region. In the case where cDNA rescue experiments work, we also test whether introduction of a mutant cDNA defective in the druggable activity (e.g., kinase dead mutant) are unable to rescue the phenotype, thereby providing evidence that small molecule inhibitors might produce similar phenotypes.
In cases where multiple targets are identified and validated, we prioritize, for further in vivo validation, those candidate targets that are (1) validated across multiple human and murine systems, (2) targetable with selective and potent small molecule inhibitors, and/or (3) abnormally expressed (up-regulated/mutated) in tumor samples corresponding to the tumor type under study. Of course, all of the screening results are considered in the context of current knowledge, and specific hits can be pursued based on a potential understanding of the underlying mechanism.
In Vivo Target Validation
Assessing the relevance of a candidate target on tumor maintenance in vivo is a critical aspect of our target validation process. A powerful feature of our pipeline for identifying and validating therapeutic targets is the ability to seamlessly take candidate results identified through in vitro screens to rapid genetic validation in genetically and pathologically accurate mouse models of human cancer. Our in vivo validation method consists of two approaches that take advantage of the inducible shRNA platform described above. First, we test whether target knockdown has a substantial impact on tumor development in tumor cell line xenograft models. Second, we incorporate ESC GEMMs that harbor multiple conditional disease-associated alleles (Premsrirut et al. 2011; Dow et al. 2012). The prototypical ESC GEMM is produced from ESCs carrying at least four alleles: (1) one or more cancer-predisposing conditional alleles that can be activated by Cre recombinase (e.g., “lox-stop-lox” KrasG12D; Tp53flox/flox); (2) a tissue-specific Cre recombinase; (3) a Cre-activatable lox-stop-lox rtTA3 allele to enable tissue-specific induction of a tet-responsive shRNA; and (4) a “homing cassette” that enables site-specific integration of the tet-responsive shRNA downstream from the Col1a1 locus using recombination-mediated cassette exchange. Other iterations omit the tissue-specific Cre (enabling somatic activation using Cre-expressing adenoviruses or lentiviruses) or link the rtTA3 allele to a fluorescent reporter (providing lineage tracing of Cre-recombined cells). For example, the CAGs-LSL-rtTA3-IRES-KATE (CAGs-RIK) transgene requires Cre activity to express the rtTA3, which is linked to the fluorescent reporter Kate and enables tissue-specific, Dox-dependent control of shRNA expression (Dow et al. 2014). These ES cells are then used to produce mice via tetraploid embryo complementation or blastocyst injection to generate multiallelic mouse models without extensive strain intercrossing.
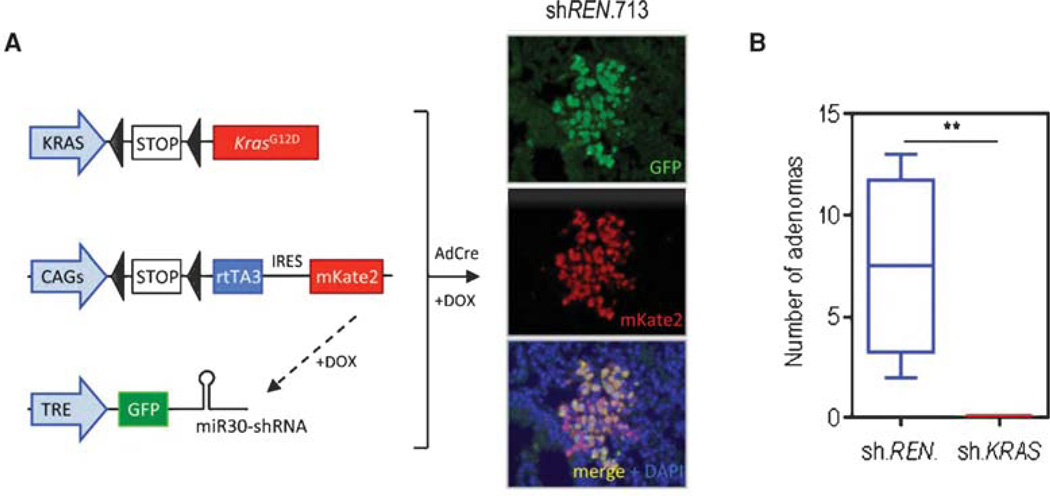
Once established, multiallelic ES cell lines can be further manipulated in vitro (to introduce additional genetic complexity), for instance, by genome editing of other disease modifying genes or through recombination of a genetic element into the homing cassette. As additional genetic alterations are introduced in culture, multiple genes can be assessed in parallel without the need for large, expensive breeding colonies. Moreover, virtually all of the derived mice have a relevant genotype and can be used in experimental studies (Premsrirut et al. 2011). In one validation of the system, we generated an ESC GEMM lung adenocarcinoma model harboring LSL-KrasG12D; CAGs-RIK; and tetracycline response element (TRE)-shKras transgenes and showed that induction of the Kras shRNA in established adenomas by doxycycline treatment (12 wk upon mutant Kras activation by intratracheal delivery of Adenovirus Cre) led to complete tumor regression compared with mice expressing a control shRNA (sh.REN) (Fig. 2). Similar results are observed in an ESC GEMM of Myc-overexpressing HCC, where induction of Cdk9 shRNAs blocked tumor progression when expressed in established liver tumors (data not shown).
Figure 2.
Embryonic stem cell (ESC)-based genetically engineered mouse models (GEMMs) for in vivo target validation. (A) Schematic representation of LSL-KrasG12D; CAGs-LSL-rtTA3-IRES-mKate (RIK); and TRE-GFP-miR30 alleles, before Cre-mediated recombination. Immunofluorescent stains for green fluorescent protein (GFP) and mKate2 in lung sections of triple transgenic mice (CAGs-LSL-rtTA3-IRES-Kate/sh-Ren.713/LSL-KrasG12D). (B) Quantification of lung adenomas in LSL-KrasG12D carrying either a control (Ren.713) or Kras-targeted short-hairpin RNA (shRNA) following 2 wk of treatment with doxycycline (**, P < 0.01). All mouse experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee (protocol no. 12-04-006). Statistical significance was calculated by two-tailed Student’s t-test (Prism 6 software). Significance values are *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
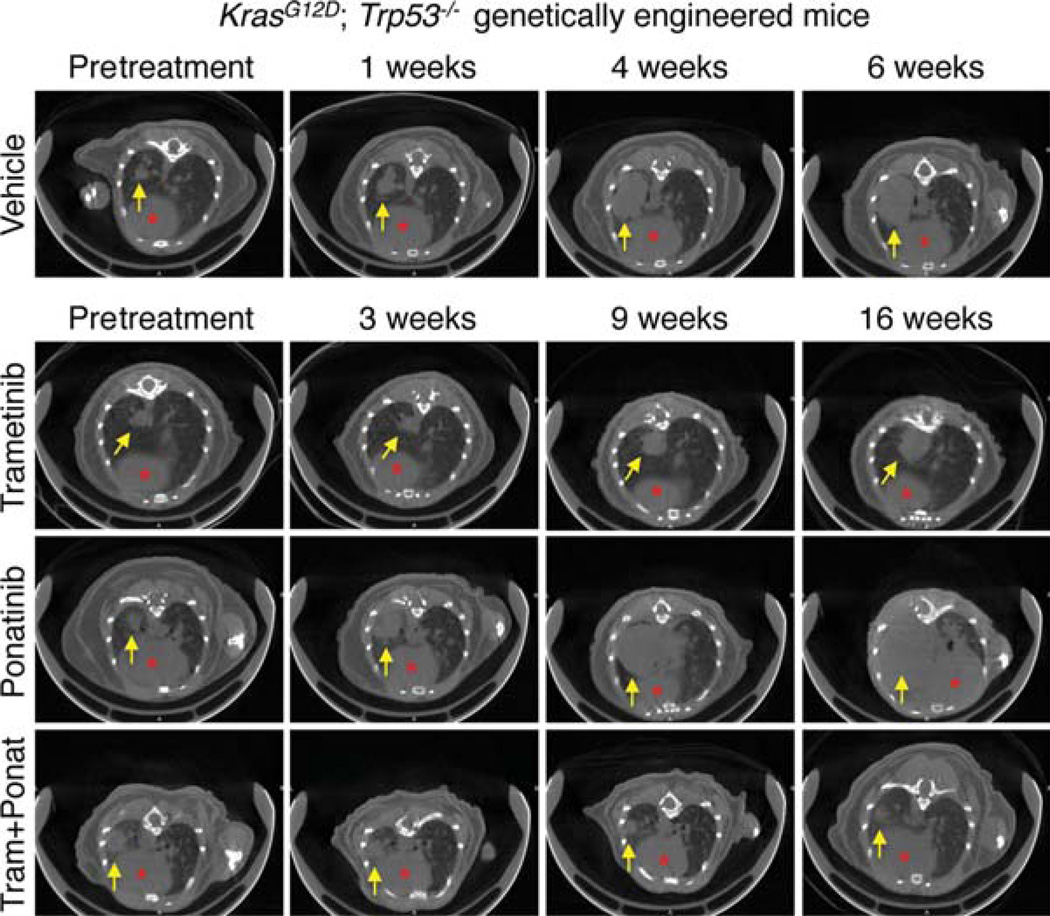
One advantage of our ESC GEMM approach is that it produces genetically and physiologically accurate preclinical models for studying the impact of small molecule regimens in established tumors. In situations where a drug against the target exists, we use the in vivo models described above to compare and contrast the impact of genetic versus small molecule action, and begin to gain insights into factors that influence de novo or acquired resistance to such pharmacological treatments. Illustrating this point, we recently validated the combined inhibition of MEK and FGFR1 as a therapeutic strategy using a GEMM of KrasG12D lung adenocarcinoma. Although trametinib and ponatinib, an FDA-approved multikinase inhibitors that inhibit FGFR1, had minor antitumor effects as single agents, the drug combination caused significant tumor regression characterized by massive necrosis (Fig. 3; Manchado et al. 2016).
Figure 3.
Ponatinib in combination with trametinib leads to tumor regression of KRAS-mutant lung tumors. Representative µCT images of the lungs of KrasG12D; Trp53−/− genetically engineered mice treated with vehicle, trametinib (3 mg/kg), ponatinib (30 mg/kg), or both drugs in combination for the indicated times. Lung tumors are indicated by yellow arrows and red asterisks mark the hearts. All mouse experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee (protocol no. 12-04-006).
The above-described ESC GEMMs enable deep and robust validation of potential cancer targets identified through shRNA screens. Of course, human-based models such as patient-derived xenograft systems are also required to establish human relevance (Manchado et al. 2016). By combining these and other orthogonal methods, we strive to capture the genetic complexity and heterogeneity of the human disease, as well as the in vivo physiological tumor environment of genetically and pathologically accurate cancer mouse models.
ESTABLISHING A THERAPEUTIC INDEX USING IN VIVO RNAi
A crucial but often overlooked step of target validation is identifying the potential toxicities that can arise from target inhibition in normal tissues. Although many of the studies that strive to identify targets through functional genomics confirm a differential sensitivity between tumor and normal cells of the same tissue, they rarely address the critical issues that may produce dose-limiting toxicities in vivo (e.g., toxicity to the hematopoietic system or the gut). All too often, the liabilities of certain targets are not identified until toxicity studies are performed on clinical candidate compounds. In some instances, these toxicities can be “off-target,” arising from a lack of drug specificity and the deleterious consequences of inhibiting an unintended target. Although such toxicities can be overcome by producing more selective agents, other toxicities are “on-target” and reflect the essential function of the target protein in certain normal tissues. Knowledge of potential “on-target” toxicities would be extremely valuable before initiating expensive drug development efforts.
Genetic studies in mouse models provide one way to reveal the function of a gene product in specific tissues or throughout the animal and, in principle, can identify pathologies associated with inhibiting a candidate drug target. In practice, these models are rarely used for this purpose. This underutilization is partially due to concerns that the consequences of gene deletion during development may not reflect the impact of acute protein inhibition in the adult. In addition, it has historically been extraordinarily time consuming and expensive to produce the appropriate models, and the relevant approaches are typically best suited for the academic setting where questions related to drug toxicity are not a priority. To address some of these limitations, we combine the optimized shRNA technology above with modeling approaches that allow acute and reversible gene suppression in normal tissues and used these to study the requirement for candidate drug targets across a wide range of adult tissues. Although recognizing that genetic studies in mice will never fully reflect the impact of drugs on their targets in normal human tissues, we believe that this approach will facilitate the assessment of on-target toxicities associated with target inhibition and modify the course of drug development. To illustrate our approach, we describe efforts using RNAi to explore the consequences of liver-specific Cdk9 suppression and systemic Brd4 suppression in vivo.
Liver Regeneration for Assessing Toxicity of Target Inhibition
As described above, the identification of CDK9 as an important maintenance gene in MYC-overexpressing cancers fueled interest in targeting transcription elongation as an anti-MYC strategy (Huang et al. 2014). CDK9, a key transcription elongation factor, plays an important role in transcriptional amplification and is involved in many essential processes. Still, there is evidence that some cells depend more on CDK9 than others, and as such it is considered not only as a promising drug target for cancer but also for diseases such as cardiac hypertrophy (Krystof et al. 2010) and HIV/AIDS (Wei et al. 1998), among others (Wang and Fischer 2008). Although several CDK9 inhibitors are available, these agents are invariably toxic (Wang and Fischer 2008), most likely because of their lack of exquisite on-target selectivity to CDK9 and frequent off-target inhibition of other members of the CDK family. As a consequence, it is not clear whether these toxicities are on- or off-target, and the requirement for CDK9 function in normal adult tissues remains unknown.
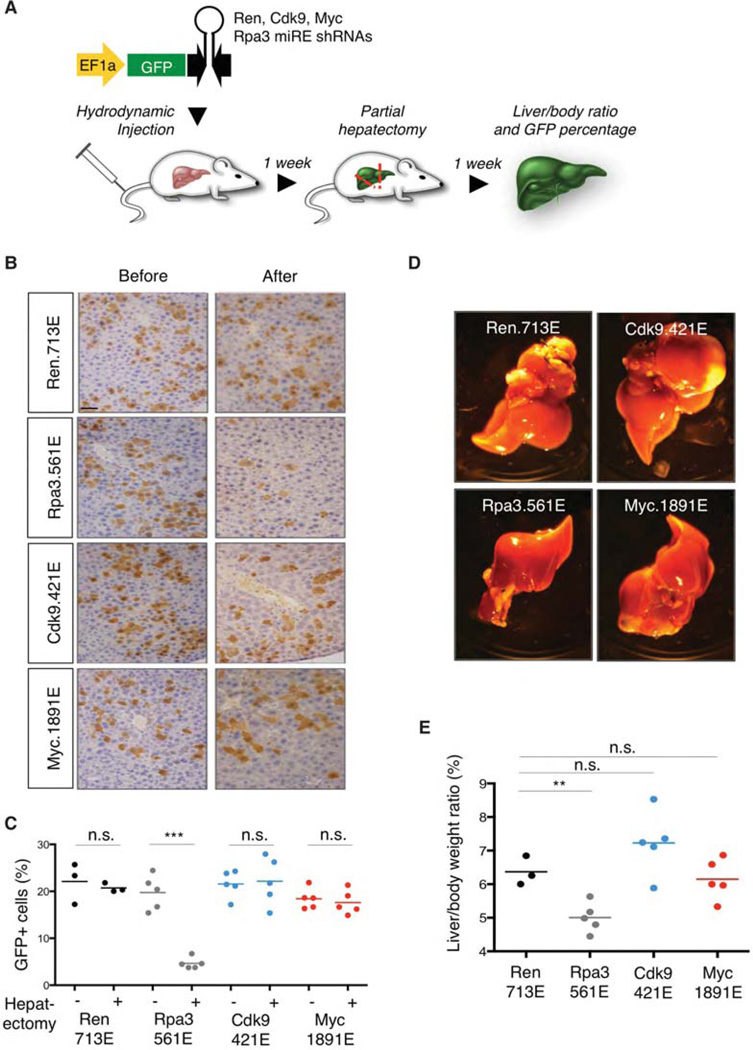
Our studies suggested that CDK9 inhibitors would be particularly effective in MYC-overexpressing HCC (Huang et al. 2014). Therefore, as a first step to assess whether CDK9 inhibition in normal hepatocytes has deleterious side effects, we performed hydrodynamic tail vein injections of transposons expressing Cdk9 or Myc shRNAs followed by partial hepatectomy (Fig. 4A). This approach leads to suppression of target genes in ~20% of hepatocytes, as revealed by GFP immunofluorescence staining (Fig. 4B,C). Suppression of replication protein A, subunit 3 (Rpa3), an essential gene in adult mice (McJunkin et al. 2011), caused about 10% reduction of green fluorescent protein (GFP)-positive hepatocytes after partial hepatectomy, which led to lower liver to body weight ratios (Fig. 4B–E). In contrast, we found that suppression of Cdk9 or Myc did not show significant effects on the proliferation of hepatocytes and liver regeneration (Fig. 4B–E) but completely abrogated Myc-driven tumor formation (Huang et al. 2014). Interestingly, a previous study also reported that conditional deletion of Myc does not impair liver regeneration (Li et al. 2006), further supporting our results using RNAi. Together, these relatively straightforward genetic experiments enabled by tracking a reporter protein in vivo provide strong evidence for a therapeutic window by directly targeting CDK9 in livers harboring MYC-overexpressing HCC.
Figure 4.
Liver regeneration for assessing toxicity of Cdk9 suppression in vivo. (A) Schematic representation of the liver regeneration. miR-E short-hairpin RNA (shRNA) transposon vectors targeting control Renilla luciferase, Cdk9, Myc, or Rpa3 were injected together with CMV-SB13 transposase by hydrodynamic tail vein injection into female C57BL/6 mice (6–8 wk of age) from Harlan Laboratories as previously described (Huang et al. 2014). Partial (2/3) hepatectomy (Taub 2004) was performed after 1 wk. Before being anesthetized with isoflurane, the animals received one subcutaneous dose of buprenorphine (0.5 mg/kg) and meloxicam (2 mg/kg). The median lobe, with its two central portions together with the left lateral lobe were mobilized, looped with a 4-0 vicryl ligation (Ethicon), and two-thirds of the liver volume were removed. Liver/body ratio and green fluorescent protein (GFP) percentage were examined after 2 wk. (B) Representative histological analysis of livers stained for GFP. Scale bar, 50 µm. (C) Cdk9 or Myc suppression does not show significant impact on the percentage of GFP+ cells before and after partial hepatectomy. (D) Representative images of livers injected with the indicated shRNAs after partial hepatectomy. (E) Cdk9 or Myc suppression does not show significant impact on liver/body ratio compared with Ren.713E (neutral control shRNA). Rpa3.561E is used as a positive control shRNA. All mouse experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee (protocol no. 11-06-011). Statistical significance was calculated by two-tailed Student’s t-test (Prism 6 software). Significance values are *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Studying the phenotypes associated with silencing CDK9 or its Cyclin partners (T1, T2, or K) systemically in vivo using genetic approaches has been challenging owing to embryonic lethality (Kohoutek 2009). Thus, further investigating the consequence of systemic Cdk9 suppression in adult mice using RNAi will be important to identify potential toxicities associated with CDK9 inhibition and understand its functions in vivo. Indeed, studies are underway to use ESC GEMMs to inducibly and systemically inhibit Cdk9 as discussed for Brd4 below.
Inducible and Reversible Transgenic RNAi Mice for Assessing Target Inhibition-Associated Toxicity
As described above, our laboratory has established an ESC GEMM platform for the production of transgenic mice harboring tetracycline/doxycycline (dox) responsive shRNAs that allows temporal and spatial control of endogenous gene expression (Premsrirut et al. 2011). Because shRNAs do not modify the target gene, this system enables reversible gene silencing (Premsrirut et al. 2011; Dow et al. 2012), thereby facilitating assessment of reversible and irreversible loss of function phenotypes. For example, we have studied the functions of Apc (Premsrirut et al. 2011; Dow et al. 2015), Rpa3 (McJunkin et al. 2011), and Pten (Miething et al. 2014) under different conditions in vivo using inducible and reversible RNAi.
More recently, we began harnessing this platform to study drug target genes identified from the RNAi screening pipeline described earlier. When applied for target validation, such system can discover potential toxicities associated with on-target gene suppression, determine the kinetics of the toxicities upon target inhibition, and examine whether any or all are reversible. To illustrate the feasibility of this approach, we generated inducible Brd4 shRNA transgenic mice to identify the consequences of Brd4 inhibition in adult tissues (Bolden et al. 2014). Suppression of Brd4 in the hematopoietic system produced defects in hematopoiesis, with an impact on the proliferation of several lineages, and a general depletion of stem cells (Bolden et al. 2014). In addition, acute Brd4 inhibition disrupted the epidermis plus hair follicles and triggered the depletion of multiple cell types in the intestine (Bolden et al. 2014). Remarkably, these phenotypes were rapidly reversible, and mice completely recovered after restoration of normal Brd4 levels. Collectively, these data suggest that “on-target” Brd4 inhibition can produce deleterious pathologies but raise hope that these might be managed by dose and schedule adjustment.
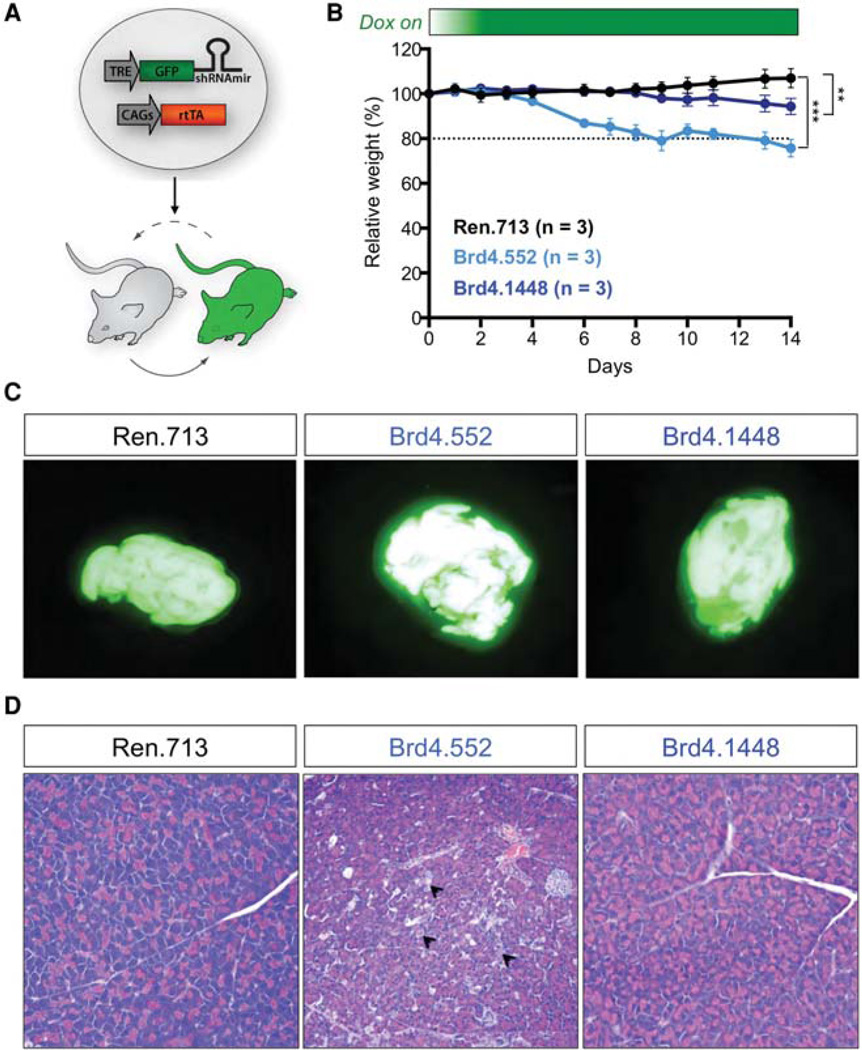
The studies described above use mice in which the GFP/Brd4 shRNA cassette was expressed using the TRE-tight promoter, which we subsequently realized limited expression of the shRNAs to only a subset of tissues (Bolden et al. 2014). In more recent efforts, we have largely eliminated these tissue restrictions by expressing the shRNA from the conventional TRE promoter. To see whether this system would identify additional pathologies for Brd4 inhibition, ESC clones were targeted with two independent GFP-linked Brd4 miR-30 shRNAs expressed downstream from the TRE promoter (hereafter referred to as TG-Brd4.552 and TGBrd4.1448) (Fig. 5A). Next, we examined the transgenic mice (CAGs-rtTA3; TG-shBrd4 mice) on a dox diet for 2 wk. Both dox-fed Brd4 shRNA mice showed significant weight loss over 2 wk (TG-Brd4.552: ~20%; TGBrd4.1448: ~6%) compared to control TG-Ren.713 mice (Fig. 5B). Of note, transgenic mice harboring Brd4 shRNAs driven by TRE-tight promoter only showed reduced weight gain (Bolden et al. 2014), suggesting additional toxicities not observed with the TRE-tight system (data not shown). By evaluating the GFP-expressing organs from TG-Brd4.552 and TG-Brd4.1448 mice and comparing to control TG-Ren.713 mice after 2 wk on dox, we identified significant lesions in the pancreas (Fig. 5C). The lesions resulted from extensive scattered acinar cells undergoing necrosis and apoptosis (Fig. 5D), suggesting that Brd4 may play an important role in pancreas homeostasis and function. Together, this systemic RNAi model identified toxicity in the pancreas, in addition to epidermal hyperplasia, alopecia, and stem cell depletion in the small intestine, as described previously (Bolden et al. 2014).
Figure 5.
Inducible and reversible transgenic RNA interference (RNAi) mice for assessing toxicities associated with systemic Brd4 suppression. (A) Schematic representation of the generation and application of short-hairpin RNA (shRNA) transgenic mice. Tetracycline response element (TRE)-driven miR-30 shRNAs are targeted to the ColA1 locus to drive doxycycline (dox)-dependent genes knockdown in embryonic stem cells and embryonic and adult tissues of the mouse as previously described (Bolden et al. 2014). By further using a sensitive and widely expressed rtTA mice strain, CAGs-rtTA3, GFP-miR-30 shRNAs can be efficiently expressed in most tissues to study on-target toxicities. (B) Mean weight changes of male and female (combined) CAGs-rtTA3/+; TG-Ren.713, TG-Brd4.552, and TG-Brd4.1448 mice on dox diet, relative to day 0 of dox treatment. Error bars represent ± SD. (C) Green fluorescent protein (GFP) expression in pancreas from CAGs-rtTA3/+; TG-Ren.713, TG-Brd4.552, and TG-Brd4.1448 mice, maintained on dox diet for 2 wk. (D) Representative hemotoxylin and eosin (H&E) stains of pancreas from CAGs-rtTA3/+; TG-Ren. 713, TG-Brd4.552, and TG-Brd4.1448 mice maintained on dox diet for 2 wk. Arrowheads indicate the extensive scattered acinar cells undergoing necrosis. All mouse experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee (protocol no. 11-06-011). Statistical significance was calculated by two-tailed Student’s t-test (Prism 6 software). Significance values are *, P < 0.05; **, P <, 0.01; and ***, P < 0.001.
The data from these Cdk9 and Brd4 studies highlight how the RNAi technology can be used to explore in vivo gene-specific toxicities, in principle, even in advance of therapeutic development, and to explore the tolerability of certain therapeutic strategies. Although RNAi-mediated protein knockdown may not always produce the same effects as small molecule protein inhibition, the systems above can examine the effects of acute, partial, and reversible consequences of target inhibition, providing a “first pass” assessment of therapeutic index to mimic phase I studies in clinical trials. Moreover, RNAi can be further used to precisely reveal the toxicities associated with recently developed phthalimide conjugation inhibitors for in vivo target protein degradation such as dBET, a bifunctional thalidomide-conjugated version of JQ1 (Winter et al. 2015). Indeed, clinical trials on various BET inhibitors are well underway, and early data suggest that hematologic and gut toxicities are dose limiting—toxicities that were predicted using our thorough RNAi target validation pipeline (Stathis et al. 2016). Still, to date, skin pathologies have not been reported. Although the basis for this discrepancy remains unknown, it is worth reiterating that the shRNA models show potent and sustained target inhibition 24/7, which may not be achieved with the clinical candidate. Regardless, these models can be effectively used to help anticipate any potential on-target toxicities that might be produced by a new agent.
CONCLUSION
In the past few years, we have made significant advances in understanding the molecular lesions that contribute to tumor formation. Although these efforts have led to the development of a number of successful molecularly targeted therapies, an increase in our molecular understanding of aberrant cancer cell signaling can lead to the functional identification of new vulnerabilities and, consequently, to more efficient therapies. RNAi, as well as emerging genome-editing technologies, represent a powerful way of revealing such cancer dependencies. The approaches described here uniquely exploit optimized RNAi screening systems for target identification as well as in vivo approaches for deep target validation. Together, these optimized RNAi approaches and additional orthogonal approaches, including new genome editing technologies, will facilitate the identification and validation of cancer targets whose further development is likely to produce real clinical benefit.
Acknowledgments
We thank S. Houlihan, M. Taylor, D. Grace, and J. Simon for excellent technical assistance and F.J. Sánchez- Rivera for carefully editing the manuscript. We also thank members of the Lowe laboratory for stimulating discussions and, in particular, Dr. Charles J. Sherr for important suggestions. We thank Memorial Sloan Kettering Cancer Center (MSKCC) Animal Facilities and MSKCC Genomics Core. E.M. was supported by The Jane Coffin Childs Memorial Fund for Medical Research and a K99/R00 grant from the National Institutes of Health (NIH)/National Cancer Institute (NCI). C.-H.H. was funded by the Ministry of Education of Taiwan. This work was supported by program project grants from the NCI (S.W.L.) and Cancer Target Discovery and Development Center (S.W.L.). S.W.L. is the Geoffrey Beene Chair for Cancer Biology and a Howard Hughes Medical Institute investigator.
REFERENCES
- Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, Clevers H, Lowe SW. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 2014;8:1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, Xuan Z, Zuber J, Wigler M, Hicks J, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Deans RM, Morgens DW, Okesli A, Pillay S, Horlbeck MA, Kampmann M, Gilbert LA, Li A, Mateo R, Smith M, et al. Parallel shRNA and CRISPR–Cas9 screens enable antiviral drug target identification. Nat Chem Biol. 2016;12:361–366. doi: 10.1038/nchembio.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA, Hannon GJ, Lowe SW. A pipeline for the generation of shRNA transgenic mice. Nat Protoc. 2012;7:374–393. doi: 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Nasr Z, Saborowski M, Ebbesen SH, Manchado E, Tasdemir N, Lee T, Pelletier J, Lowe SW. Conditional reverse tet-transactivator mouse strains for the efficient induction of TRE-regulated transgenes in mice. PLoS One. 2014;9:e95236. doi: 10.1371/journal.pone.0095236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, Lowe SW. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161:1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ, Kitzing T, Zhu N, de Stanchina E, Sawyers CL, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28:1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohoutek J. P-TEFb—The final frontier. Cell Div. 2009;4:19. doi: 10.1186/1747-1028-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystof V, Chamrad I, Jorda R, Kohoutek J. Pharmacological targeting of CDK9 in cardiac hypertrophy. Med Res Rev. 2010;30:646–666. doi: 10.1002/med.20172. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xiang Y, Potter J, Dinavahi R, Dang CV, Lee LA. Conditional deletion of c-myc does not impair liver regeneration. Cancer Res. 2006;66:5608–5612. doi: 10.1158/0008-5472.CAN-05-4242. [DOI] [PubMed] [Google Scholar]
- Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–290. [PubMed] [Google Scholar]
- Manchado E, Weissmueller S, Morris JP, 4th, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. A combinatoria l strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, Stillman B, Lowe SW. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci. 2011;108:7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miething C, Scuoppo C, Bosbach B, Appelmann I, Nakitandwe J, Ma J, Wu G, Lintault L, Auer M, Premsrirut PK, et al. PTEN action in leukaemia dictated by the tissue microenvironment. Nature. 2014;510:402–406. doi: 10.1038/nature13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord JP, de La Motte Rouge T, Uro-Coste E, de Braud F, Pelosi G, French CA. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R. Liver regeneration: From myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- Wang S, Fischer PM. Cyclin-dependent kinase 9: A key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharmacol Sci. 2008;29:302–313. doi: 10.1016/j.tips.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Watanabe C, Cuellar TL, Haley B. Quantitative evaluation of first, second, and third generation hairpin systems reveals the limit of mammalian vector-based RNAi. RNA Biol. 2016;13:25–33. doi: 10.1080/15476286.2015.1128062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Weissmueller S, Manchado E, Saborowski M, Morris JP, 4th, Wagenblast E, Davis CA, Moon SH, Pfister NT, Tschaharganeh DF, Kitzing T, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell. 2014;157:382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011a;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011b;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]