Fig. 5.
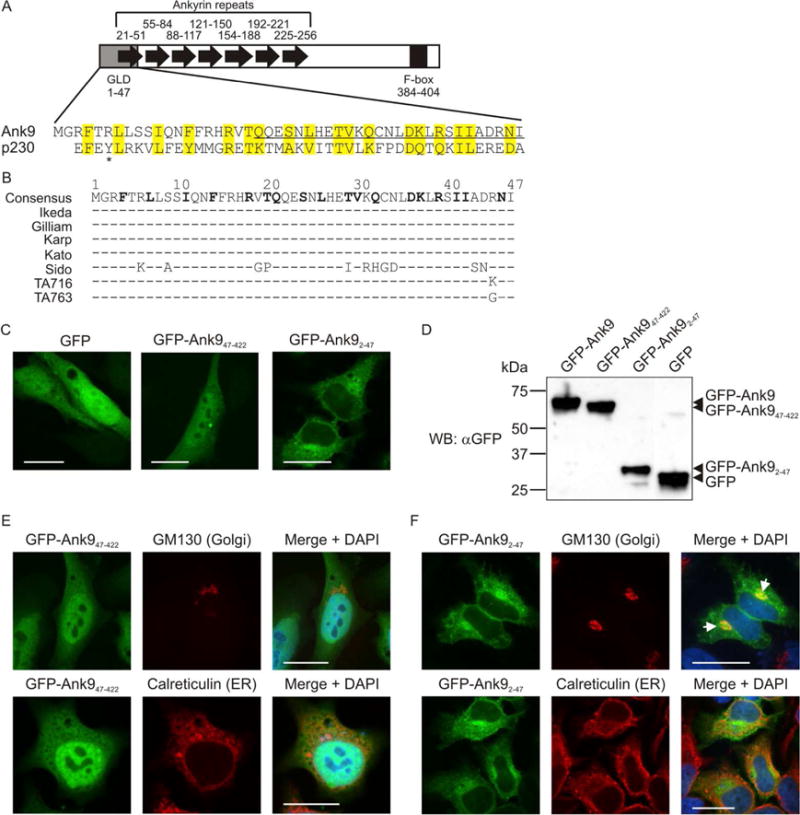
The conserved Ank9 N-terminus is both necessary and sufficient for Golgi localization, but is insufficient for Golgi-to-ER retrograde trafficking. (A) Schematic of Ank9. A gray rectangle denotes the Golgi localization domain (GLD). Black arrows denote individual ankyrin repeats. A black rectangle denotes the F-box. Numbers above or below each symbol represent the amino acids that make up the cognate motif. The Ank9 N-terminal GLD (amino acids 1–47) bears sequence similarity to the eukaryotic p230 GRIP domain, as shown in the amino acid alignment. Identical or similar amino acid residues are highlighted in yellow. An asterisk denotes the Y residue at position 4 of p230 that is conserved among eukaryotic GRIP domains. (B) Conservation of the Ank9 N-terminal GLD among O. tsutsugamushi strains. An alignment of amino acids 1 through 47 from seven O. tsutsugamushi Ank9 homologs is shown. Strain names are listed to the left of each sequence. Amino acid positions are denoted with numbers above the consensus sequence. Consensus sequence residues in bold text have identity/similarity to those of the p230 GRIP domain as shown in panel A. For each O. tsutsugamushi strain sequence, residues identical to the consensus sequence are represented by a dash (−) while non-identical amino acids are shown as their corresponding single letters. (C, E, and F) Ank9 residues 1–47 are sufficient and necessary for GFP-Ank9 to localize to the Golgi but are insufficient for GFP-Ank9 to retrograde traffic from the Golgi to the ER. HeLa cells transfected to express GFP, GFP-Ank947–422 (lacks the GLD), or GFP-Ank92–47 (GLD alone) were fixed and analyzed by confocal microscopy (C) or screened with antibody against the Golgi marker, GM130, or the ER marker, calreticulin, prior to confocal microscopic examination (E and F). Representative fluorescence images of cells viewed for GFP, GM130 or calreticulin, and merged images plus DAPI are presented. (E and F) Representative cells exhibiting GFP-Ank9 Golgi-associated (Golgi) and ER-associated (ER) subcellular localization patterns are denoted. White arrows in F designate representative points of GFP and GM130 signal colocalization. Scale bars, 20 μm. (D) Confirmation of expected protein sizes for GFP-Ank9, GFPAnk947–422, GFP-Ank92–47, and GFP. HeLa cell lysates of cells expressing GFP fusion proteins were analyzed by Western blot with GFP antibody. Arrowheads mark the expected apparent molecular weights for noted proteins. Results shown are representative of three independent experiments.
