Figure 2.
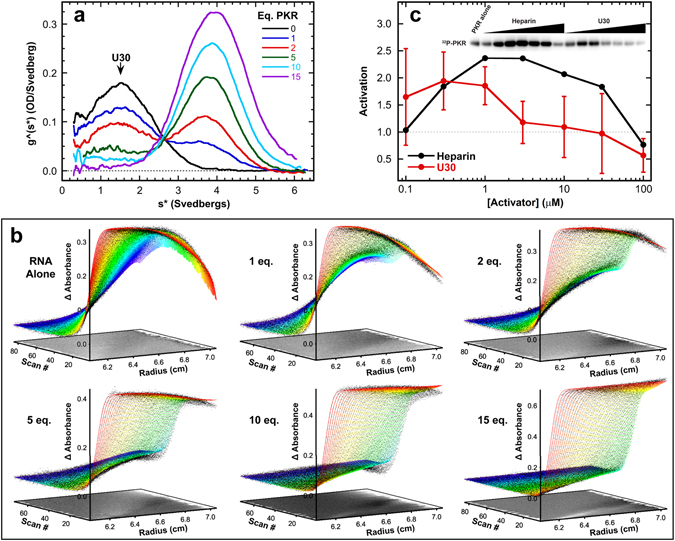
Interaction of PKR with ssRNA. PKR binding to U30 ssRNA was assayed by sedimentation velocity analytical ultracentrifugation. Measurements were performed in AU75 buffer at 20 °C and 50,000 rpm using absorbance detection at 260 nm. (a) Titration of U30 with PKR represented as an overlay of g^(s*) sedimentation coefficient distribution functions. The samples contained 1 µM U30 ( ) and 1 µM U30 plus 1 eq. (
) and 1 µM U30 plus 1 eq. ( ), 2 eq. (
), 2 eq. ( ), 5 eq. (
), 5 eq. ( ), 10 eq. (
), 10 eq. ( ), and 15 eq. PKR (
), and 15 eq. PKR ( ). The decrease in the U30 peak and appearance of the peak at higher S are due to complex formation. (b) Global analysis of the time difference curves. Scans within each data set were subtracted in pairs to remove time-invariant background noise and fit to a sequential 2:1 binding model using SEDANAL53. The data are indicated by points and the fit by solid lines. The residuals are plotted as a grayscale image in the x-y plane at z = 0. The best-fit parameters are in Table 1. (c) Activation of PKR by U30. 500 nM PKR was incubated with variable concentrations of U30 in AU75 buffer with 5 mM MgCl2 for 20 min at 32 °C. Samples were resolved by SDS-PAGE and 32P-PKR was quantified with a phosphorimager. The data are normalized to activation of PKR in the absence of activator. The error bars correspond to the standard deviation from three replicates. The inset shows a cropped image of the gel.
). The decrease in the U30 peak and appearance of the peak at higher S are due to complex formation. (b) Global analysis of the time difference curves. Scans within each data set were subtracted in pairs to remove time-invariant background noise and fit to a sequential 2:1 binding model using SEDANAL53. The data are indicated by points and the fit by solid lines. The residuals are plotted as a grayscale image in the x-y plane at z = 0. The best-fit parameters are in Table 1. (c) Activation of PKR by U30. 500 nM PKR was incubated with variable concentrations of U30 in AU75 buffer with 5 mM MgCl2 for 20 min at 32 °C. Samples were resolved by SDS-PAGE and 32P-PKR was quantified with a phosphorimager. The data are normalized to activation of PKR in the absence of activator. The error bars correspond to the standard deviation from three replicates. The inset shows a cropped image of the gel.
