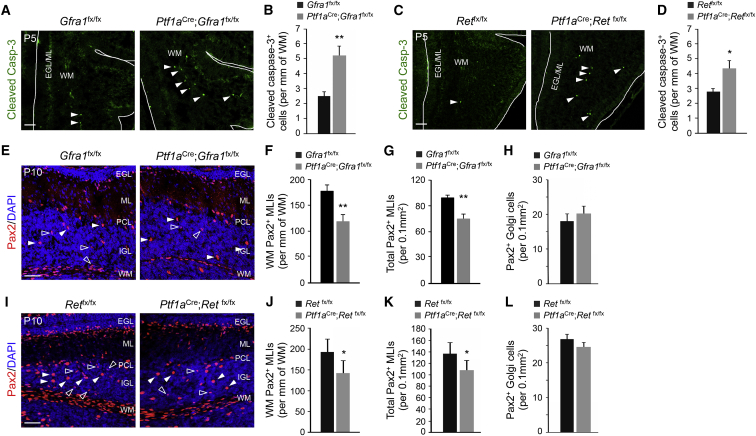
Figure 2.
Increased Apoptosis in MLI Progenitors Lacking GFRα1 or RET
(A and C) Cerebellar sections from P5 Gfra1 (A) or Ret (C) mutants and controls stained with cleaved-caspase-3 (Casp3) antibody. Arrows point to cleaved-Casp3+ cells in the WM. White lines delineate the border of the folium. EGL, external granule layer; ML, molecular layer; WM, white matter. The scale bar represents 50 μm.
(B and D) Quantification of cleaved-Casp3+ cells in the folial WM of control and Gfra1 (B) or Ret (D) mutants. Values are mean ± SEM. n = 6 mice per group.
(E and I) Cerebellar sections from P10 Gfra1 (E) or Ret (I) mutants and controls stained for Pax2 and counterstained with DAPI. Migratory MLIs (open arrows) and Golgi cells (solid arrows) are indicated. PCL, Purkinje cell layer; IGL, internal granule layer. The scale bar represents 50 μm.
(F, G, J, and K) Quantification of folial WM (F and J) or total (G and K) Pax2+ MLI progenitor densities in P10 Gfra1 (F and G) or Ret (J and K) mutants and controls. Values are mean ± SEM. n = 6 (F and G) or 5 (J and K) mice per group.
(H and L) Quantification of Pax2+ Golgi cells in the IGL of P10 Gfra1 (H) or Ret (L) mutants and controls. Values are mean ± SEM. n = 6 and 5 or 8 and 6 mice per group, respectively.