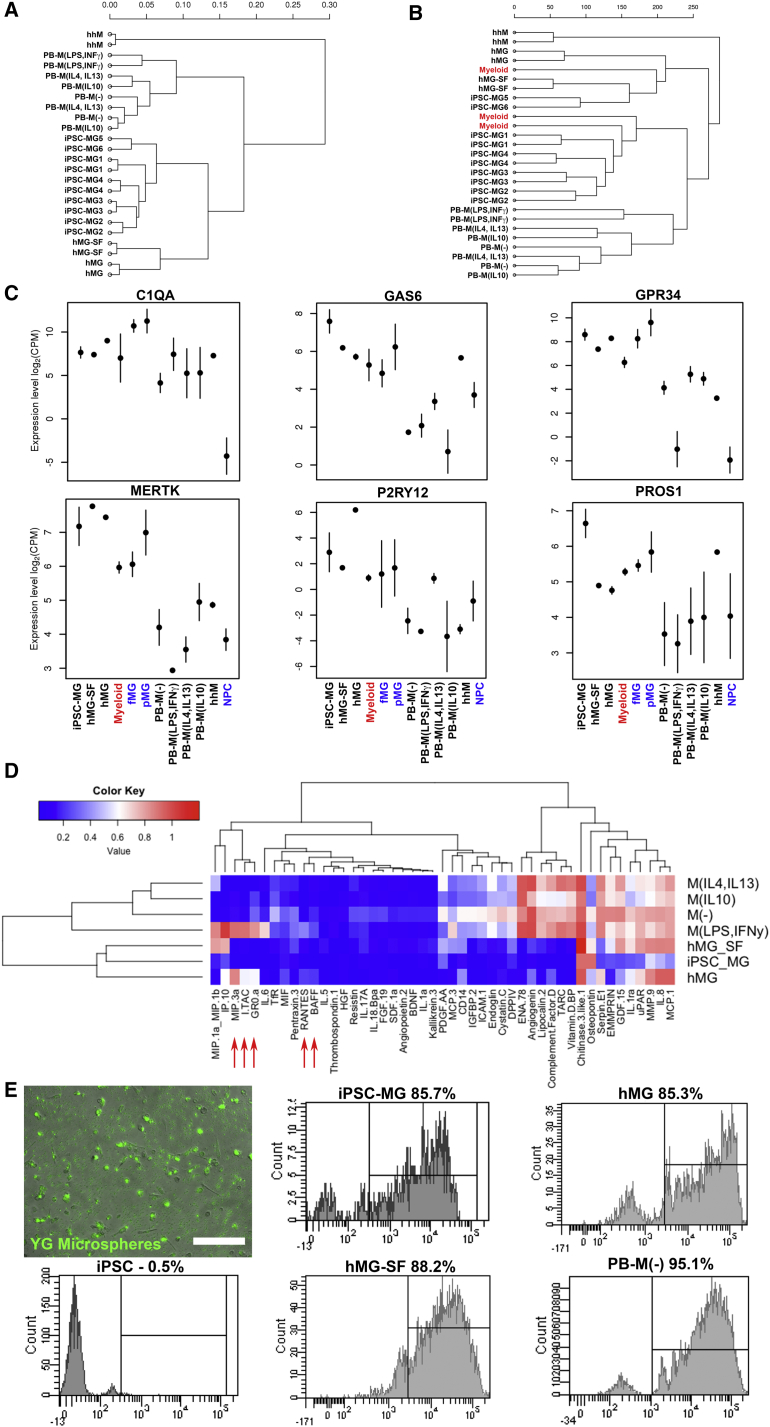
Figure 3.
Gene Expression, Cytokine Release Profile, and Phagocytosis of Microglia and Macrophages
(A) Hierarchical clustering dendrogram of the RNA-seq data based on global mRNA expression. Sample distances were calculated from Pearson's correlation coefficient.
(B) Dendrogram showing hierarchical clustering of our RNA-seq data and data obtained from an independent study of human primary CD45+ cells in the brain (“Myeloid” samples in red, GEO: GSE73721). Analysis is based on transcriptome-wide expression.
(C) Graphs showing the expression levels of the six human microglial signature genes. Error bars are means ± SD. Colored samples correspond to data from independent studies (GEO: GSE73721 in red; GEO: GSE85839 in blue).
(D) Heatmap of the released cytokine profiles of five independent iPSC-MG runs from two lines, two independent hMG samples, and one hMG-SF sample compared with PB-M. Red arrows indicate the five proteins upregulated in hMG and M(LPS,IFNγ).
(E) Representative fluorescent image and flow-cytometry histograms showing phagocytosis of yellow-green (YG)-labeled microspheres. Scale bar, 200 μm. iPSC, undifferentiated iPSCs used as negative control.