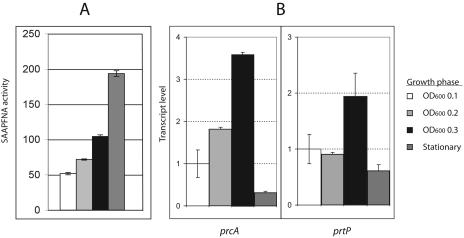
FIG. 1.
Protease activity and transcription of prcA-prtP in T. denticola 35405. (A) SAAPFNA activity (8) expressed in A405/h/ml/OD600. Error bars represent the range of values obtained with triplicate samples of a representative experiment. (B) QRT-PCR of prcA and prtP. Total RNA was extracted from cultures harvested at an OD600 of 0.1, 0.2, and 0.3 and at stationary phase (fifth day). DNase-treated RNA samples were reversed transcribed with random hexamer primers by using the SuperScript First-Strand synthesis system for RT-PCR (Invitrogen). One microliter of the resulting first-strand cDNA was amplified by using QuantiTect SYBR Green PCR (QIAGEN) in 25 μl of reaction buffer, with 16S rRNA serving as an internal control for normalization between samples. Gene-specific primers separated by approximately 80 to 100 bp were designed with Primer Express software (Perkin-Elmer Applied Biosystems). Thermal cycling was performed in an iCycler iQ Multi-Color Real Time PCR detection system (Bio-Rad) at 95°C for 15 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. Threshold values were calculated by using baseline cycles 2 to 10. Data were analyzed by using software supplied by the manufacturer. The data are shown with the gene expression level defined as 1.0 at an OD600 of 0.1. Error bars represent standard deviations of the means of results from triplicate samples in two independent experiments.