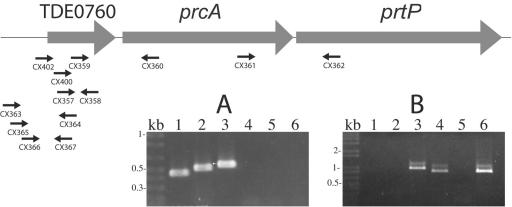
FIG. 3.
RT-PCR analysis of prcA-prtP operon. DNase-treated RNA samples were reversed transcribed as described in the legend to Fig. 1 by using a gene-specific primer or random hexamer primers supplied with the SuperScript First-Strand synthesis system for RT-PCR (Invitrogen). One microliter of the resulting first-strand cDNA was amplified according to the manufacturer's instructions. The location and orientation of oligonucleotide primers used for RT-PCR are shown relative to TDE0760, prcA, and prtP in the graphic map. RT-PCR products, including positive (genomic DNA template) and negative (no RT enzyme) controls were analyzed by agarose gel electrophoresis. Panel A demonstrates continuity of transcription between TDE0760 and prtP. Lane 1, CX357-CX358; lane 2, CX359-CX360; lane 3, CX361-CX362; lane 4, CX363-CX364; lane 5, CX365-CX364; and lane 6, CX366-CX367. Panel B localizes the 5′ end of the transcript. CX402-CX360, lanes 1 to 3; CX400-CX360, lanes 4 to 6. Lanes 1 and 4, cDNA template; lanes 2 and 5, RNA template; lanes 3 and 6, DNA template.