Figure 1. Creation and Validation of Autophagy-Deficient Human Fibroblasts.
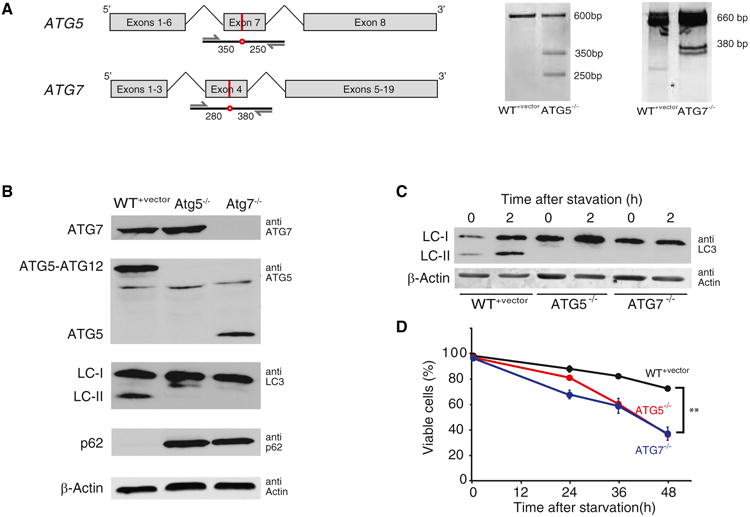
(A) The schematics on the left are gene maps of human ATG5 and ATG7 showing the site of the introduced mutations (red line and circle) and the PCR strategy for amplifying the mutated genomic regions for the SURVEYOR assay. The numbers indicate the expected sizes of the mutated amplicon fragments after SURVEYOR nuclease cleavage. The right panel shows the results of the SURVEYOR assay indicating the introduction of mutations at expected genomic positions.
(B)Western blots show the expression of ATG5, ATG7, LC3, p62, and β-actin control in WT+vector,ATG5−/−, and ATG7−/− cells. The blots indicate that ATG5 and ATG7 are completely knocked out in HCA2-hTert cells. The deletion of ATG5 and ATG7 results in complete disappearance of LC3-II, and the deletion of ATG7 prevents the conjugation of ATG5 to ATG12. The basal level of p62 is significantly elevated in ATG5−/− and ATG7−/− cells.
(C) LC3-II levels increase under starvation conditions in WT+vector, but not ATG5−/− and ATG7−/− cells.
(D) ATG5−/− and ATG7−/− cells are more sensitive to amino acid starvation. Cell viabilities were measured after different periods of amino acid starvation. Each time point was measured in three replicate experiments, and the error bar indicates SD. n = 3 biological replicates; **p < 0.01. See also Figure S1.
