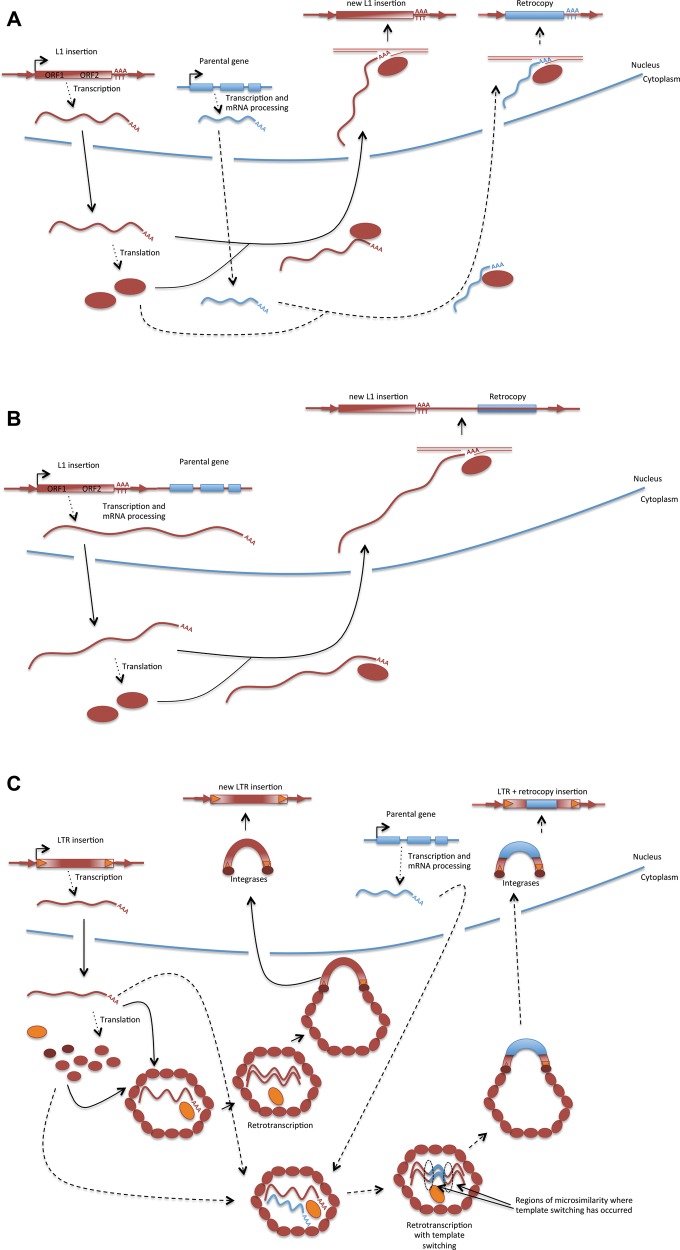
Fig. 1.
—(A) LINEs are autonomous non-LTR retrotransposable elements. They are transcribed by RNA polymerase II and their transcript encodes for a retrotranscriptase (RT) and additional activities (RNA binding and endonuclease activity within others; some not well characterized) in two open reading frames (Cost et al. 2002). The poly(A) of the L1 transcripts is bound by its proteins in the cytoplasm after the transcript is translated and carried to the nucleus. In the nucleus, the transcript undergoes target-primed reverse transcription (TPRT) after the endonuclease nicks the DNA and a 3′ end is available to prime the RT reaction (Cost et al. 2002). It is still unclear how all the reactions occur (Mandal and Kazazian 2016) but there are some clear hallmarks of L1-mediated retrotransposition: a short remnant of the poly(A) tail at the 3′ and target side duplications (TSDs; Vanin 1985). TPRT often produces 5′ truncated copies but well preserved 3′ end of the element including the short remnant poly(A) tail (Zingler et al. 2005). L1 elements can mediate the retrotransposition of mRNAs in cells. The poly(A) of cytoplasmic mRNAs might be recognized by L1 proteins, carried to the nucleus, undergo TPRT and be inserted in the genome. The hallmarks of this process are going to be the presence of an intronless copy of a gene with a remnant of a poly(A) tail at the 3′ end and TSDs flanking the insertion. 3′-UTRs will often be complete while there might be 5′ truncated copies from the onset (often the 5′-UTR can be shorter and sometimes the CDS can be affected by this truncation likely producing a retropseudogene). (B) L1 can produce the transduction of downstream regions when the transcript produced by the element is unusually long (i.e., it is not polyadenylated at the typical polyadenylation site) and includes the downstream region that could potentially encompass a gene (Goodier et al. 2000; Pickeral et al. 2000). (C) Autonomous LTR retrotransposable elements are transcribed and translated and the proteins encoded assemble a viral-like particle where reverse transcription of the LTR RNA occurs. The product will be a double-stranded LTR retroelement flanked by LTRs that proteins bind, bring to the nucleus and integrate in the genome (Levin and Moran 2011). When the reverse transcriptase switches transcripts might start retrotranscribing templates of cellular mRNAs and as it switches again to retrotranscribed the end of the LTR transcript produces a retrocopy that will often be an incomplete CDS but can potentially contain a whole retrocopy of cellular genes (Derr et al. 1991; Tan et al. 2016). The template switching will occur if there are by chance regions of sequence similarity between the mRNA and the nascent cDNA (Derr et al. 1991). When the template switching occurs more than once, chimeric retrocopies can be produced (Wang et al. 2006; Tan et al. 2016). These retrocopies or more often partial retrocopies will be flanked by LTR sequences on both sides.