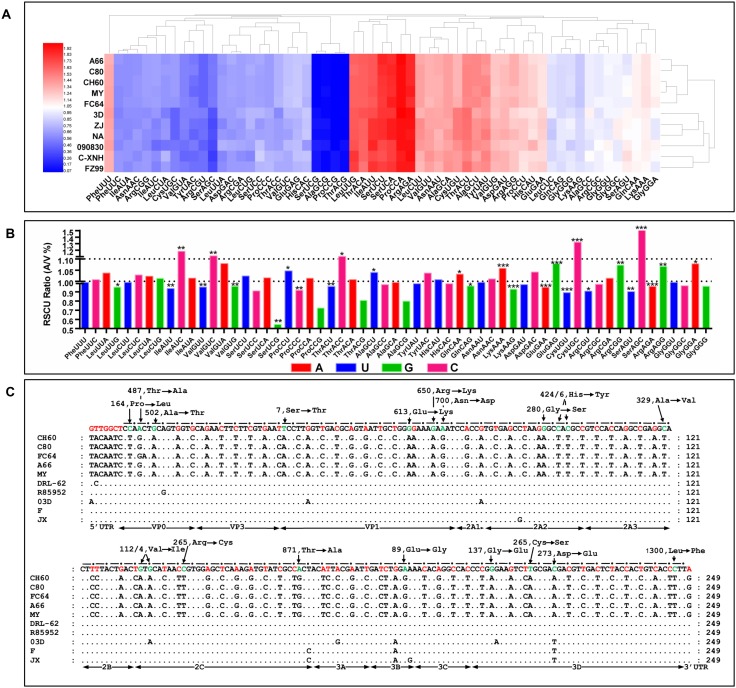
Fig 1. Codon usage bias analysis and fixed SNPs in attenuated and virulent strains.
A) RSCUs used in virulent and chick embryo attenuated strains were subject to hierarchical cluster analysis according to their Euclidean distances (Hemi software). The results indicated that the RSCU was apparently changed in the process of passage in chick embryos. B) The significance of differences of RSCU between the chick embryo attenuated strains and the virulent strains were analyzed by Student-t test. The ratios (A/V) at 1.0 and 1.1 are displayed as two dash lines. *P < 0.05, **P < 0.01 or ***P < 0.001 indicate the level of statistical significance of the differences between different groups. C) Fixed SNPs in the genomes of virulent strains and chick embryo attenuated strains. The conserved nucleotides used by virulent strains are listed on the first line, which corresponds to the dark spots. Red color indicated synonymous mutations or mutations in UTR. Green color indicated missense mutations, which are indicated at each site.