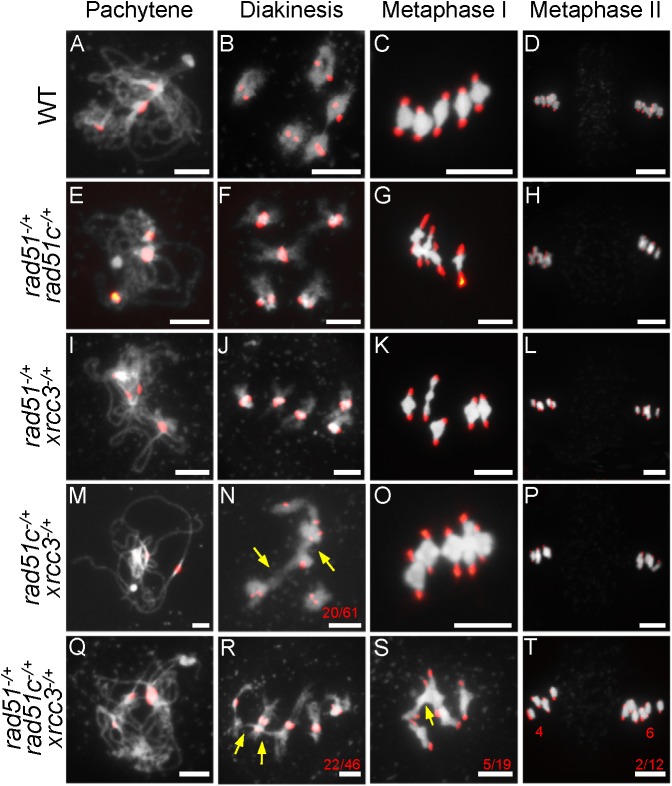
Fig 4. Centromere-fluorescence in situ hybridization analysis of chromosome morphology in double and triple heterozygous mutants.
(A-L) Wild type (WT), atrad51-/+ atrad51c-/+ and atrad51-/+ atxrcc3-/+ mutant chromosome morphologies at pachytene, diakinesis, metaphase I and metaphase II. Compared with WT, atrad51-/+ atrad51c-/+ and atrad51-/+ atxrcc3-/+ mutants had similar chromosome morphologies. (M) atrad51c-/+ atxrcc3-/+ chromosome morphology at pachytene was similar to WT, but showed 32.8% (n = 61) non-homolog association at diakinesis (N). (O, P) atrad51c-/+ atxrcc3-/+ chromosome morphology at metaphase I and II, respectively. Chromosome morphology of atrad51-/+ atrad51c-/+ atxrcc3-/+ at pachytene (Q) and diakinesis (R), metaphase I (S) and metaphase II (T). The atrad51-/+ atrad51c-/+ atxrcc3-/+ triple chromosome morphology showed more severe defects than atrad51c-/+ atxrcc3-/+. Red dots indicate centromere signals. Yellow arrows show non-homologous chromosome associations (N, R, S). The red numbers in N, R, S refer to the number of abnormal cells out of all cells observed. Scale bar: 5 μm.