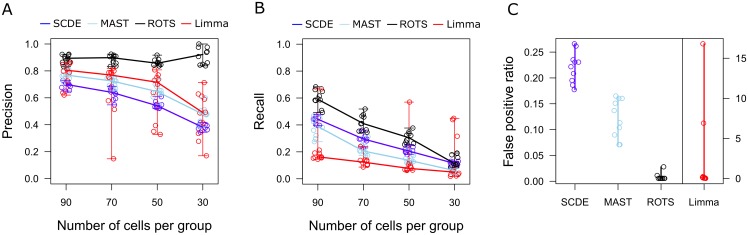
Fig 4. Precision, recall, and false positive ratios of ROTS and current state-of-the-art methods for single-cell RNA-seq in the innate lymphoid cell data.
(A) Precision of the findings in reduced data. Precision was defined as the ratio between the number of common detections in the reduced and full data, and the total number of detections in the reduced data. Median values over ten randomly generated subsets are indicated by lines across the different numbers of cells per group. (B) Recall of the findings in reduced data. Recall was defined as the ratio between the number of common detections in the reduced and full data, and the total number of detections in the full data. Median values over ten randomly generated subsets are indicated by lines across the different numbers of cells per group. (C) False positive ratios of the findings in ten randomly generated mock datasets. The false positive ratio was defined as the ratio between the number of differentially expressed genes in the mock comparison and the average number of differentially expressed genes in the actual comparison. Limma was visualized separately because of the different scale compared to the other methods and jittering was used to separate overlapping points.