Fig. 2. Association of JARID2/JMJD3 differential IR with temperature-dependent sex.
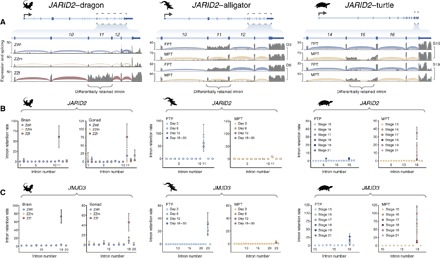
(A) Annotated gene models for the predicted ortholog of JARID2 in dragon (left), alligator (middle), and turtle (right). For dragon, normalized coverage by mapped RNA sequencing reads (gray) and density of spliced read junctions spanning annotated introns are shown for a single replicate of ZWf (blue), ZZm (yellow), and ZZf brain (red). For alligator, a single replicate of embryonic gonad after 3 and 6 days of incubation at FPT (blue) or MPT (yellow) is shown. For turtle, a single replicate of embryonic gonad at developmental stages 16 and 19 at FPT (blue) or MPT (yellow) is shown. Note that the section of zero coverage in the center of the retained intron for turtle is a string of undefined (N) bases in the genome, to which reads cannot be mapped. (B and C) Normalized rates of IR (retained transcript fragments/kilobase (kb)/spliced intron junctions; mean ± SD) are shown for individual introns in JARID2 (B) and JMJD3 (C) in ZWf, ZZm, and ZWf dragons (brain and gonad; n = 2), alligator embryonic gonad (FPT and MPT; days 3, 6, 12, and 18 to 30; n = 3), and turtle embryonic gonad (FPT and MPT; stages 15 to 19 and 21; n = 2).
