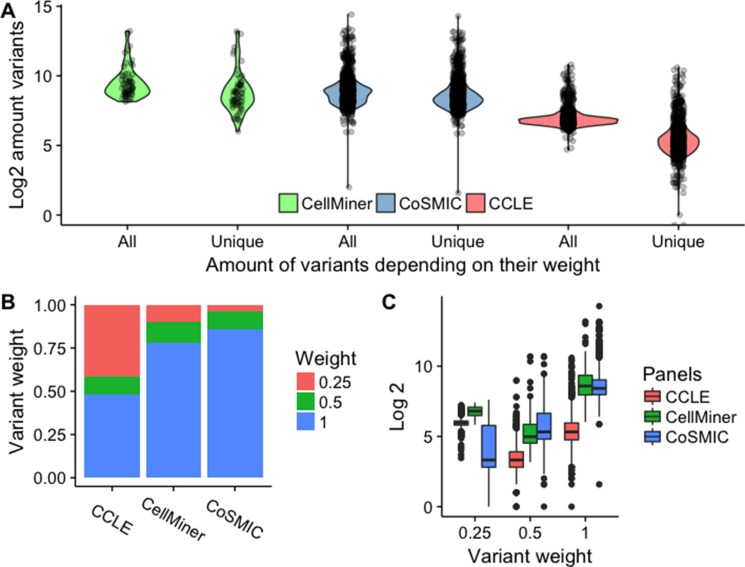
Figure 2. Distribution of CCL variant frequencies and weights across libraries.
(A) Number of “rare” variants in CCLs according to Uniquorn's weighting scheme. ‘All‘ shows the log-amount of variants per CCL without any filtering (weight 0.0) and ‘Unique shows the amount of variants that remain after all variants were filtered that were present in more than a single CCL (weight 1.0). Differences between software, technologies and filters (non-exhaustive) i.e. heterogeneous data-processing leads to different amounts of filtered, non-unique mutations as shown by the significantly different reduction of variants between the CellMiner (medium), COSMIC CLP (low) and CCLE panel (strong), see Table 3 for the sources of heterogeneity. It is shown, that all panels possess unique, i.e. ‘rare’ variants on which the Uniquorn identification method is based. (B) Distribution of weights per library. At least 50% of variants are high-weight (rare) variants. CCLE shows significantly less unique variants than COSMIC CLP and CellMiner, which explains the strong difference between raw and filtered variants in Figure A. (C) Number of variants per reference sample for different weight thresholds in the different reference libraries. CCLs from COSMIC CLP show a high amount of unique variants on average, especially when compared to those from CCLE.