Figure 2.
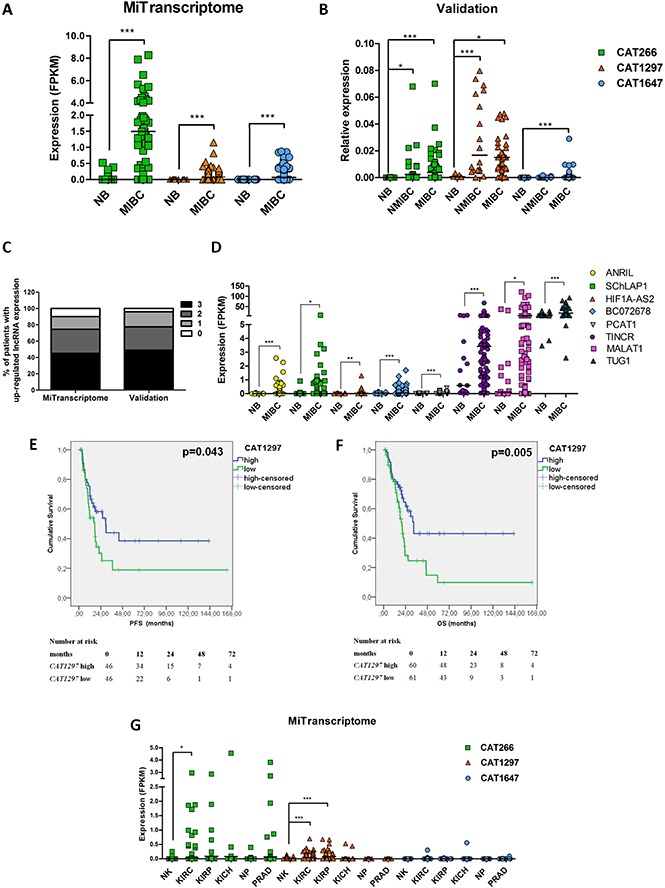
Expression of CAT266, CAT1297 and CAT1647 in normal urothelium (NB), non-muscle-invasive (NMIBC) and muscle-invasive bladder cancer (MIBC) in (A) MiTranscriptome discovery set and (B) in the Nijmegen validation set. Expression data were analyzed using unpaired t-test (*p < 0.05; **p < 0.01; ***p < 0.001, line represent the median). (C) Percentage of MIBC patients with 3, 2, 1, 0 up-regulated lncRNA in the MiTranscriptome discovery set and in the validation set in comparison to median expression levels of each lncRNA in normal urothelium. (D) Expression levels of lncRNAs described as up-regulated in bladder cancer in literature, in the MiTranscriptome cohort of MIBC. Expression data were analyzed using unpaired t-test (*p < 0.05; **p < 0.01; ***p < 0.001, line represent the median). (E) Kaplan-Meier analysis, using data from the MiTranscriptome cohort, showing correlation of CAT1297 expression levels (high n = 46; low n = 46) with progression-free survival (PFS) and (F) overall survival (OS; high n = 60; low n = 61). (G) The expression levels of CAT266, CAT1297 and CAT1647 in other urological tissue types: normal kidney (NK, n = 119), kidney renal clear cell carcinoma (KIRC, n = 463), kidney renal papillary cell carcinoma (KIRP, n = 77), kidney chromophobe renal cell carcinoma (KICH, n = 64), normal prostate (NP, n = 43) and prostate adenocarcinoma (PRAD, n = 174) in the MiTranscriptome dataset. Expression data were analyzed using unpaired t-test (*p < 0.05; **p < 0.01; ***p < 0.001, line represent the median).
