Figure 5.
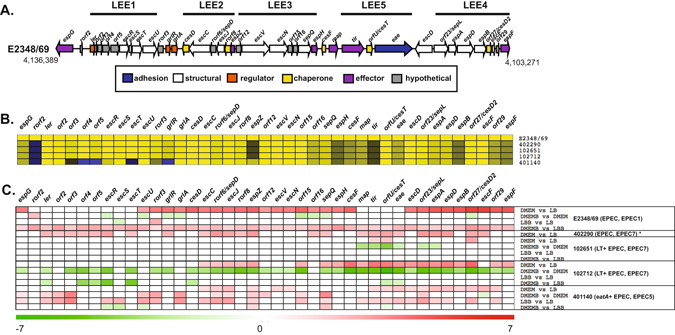
Differential expression of protein-coding genes within the LEE region. (A) Diagram of the protein-encoding genes within the LEE region of EPEC isolate E2348/69. The predicted protein function is indicated by the color of the arrow, and the size and direction of the arrows indicates the size of each predicted gene and the direction of transcription. (B) In silico detection of the protein-encoding genes of the LEE region of EPEC isolate E2348/69 in each of the EPEC genomes analyzed in this study. The colors of the heatmap represent the BSR values of each gene, with yellow indicating a gene is present, blue indicating a gene is absent, and grey to black indicating sequence divergence. (C) Heatmap of the differential expression for each of the sample comparisons of the EPEC isolates (E2348/69 and 402290) and the EPEC/ETEC hybrid isolates (102651, 102712, and 401140). Only significant LFC values ≥1 or ≤−1 are represented in the heatmap. Red indicates increased differential expression while green indicates decreased differential expression. White indicates that a gene was either not present in the EPEC isolate, and/or did not exhibit significant differential expression. *The differential expression of EPEC isolate 402290 was previously investigated during growth in DMEM vs. LB (Hazen et al. unpublished) and was included as a reference comparison in the current study; however, this EPEC isolate was not grown in LB and DMEM with added bile.
