Figure 3.
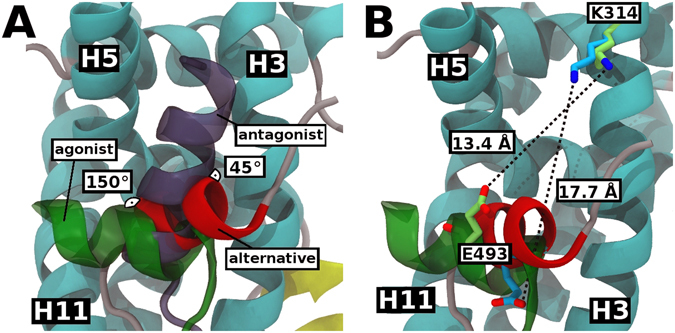
Comparison of the alternative, canonical agonist and antagonist bound structures: (A) Angle displacement between H12 position of alternative ERβ LBD-E2 (red), canonical agonist ERβ LBD-E2 (green, PDB code 3OLS) and antagonist ERβ LBD-genistein (purple, PDB code 1QKM) structures. Alternative structure appears to be in a similar position to the antagonist structure. (B) Distance between the charge-clamp residues (K314 and E493) of alternative ERβ LBD-E2 (H12 in red, residues in cyan) and canonical agonist ERβ LBD-E2 (H12 and residues in green, PDB code 3OLS). The charge clamp of canonical conformation is an optimal distance, since the structure was associated with a coactivator peptide.
