FIGURE 1.
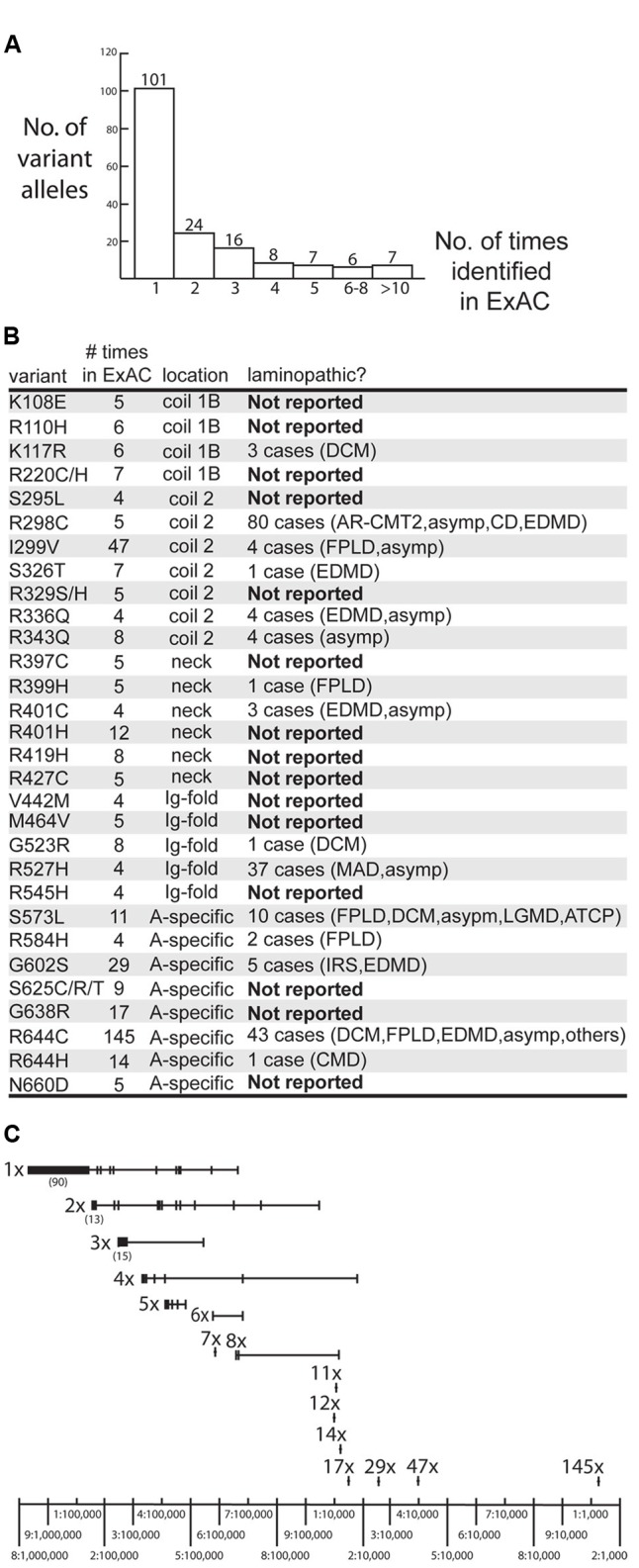
Overview of LMNA missense variants identified in the ExAC cohort. (A) Graph depicting the number of times each variant allele was identified in the ExAC cohort. Among the 169 identified variants, 101 were each identified in a single individual. (B) Information about specific variants identified four or more times in the ExAC cohort, including the number of times each variant was identified in ExAC, its molecular location in the lamin polypeptide (‘location’), and whether it is novel (‘Not Reported’), defined as not reported in the laminopathy database (comprising patients and relatives with the same LMNA mutation). AR-CMT2, autosomal recessive Charcot-Marie-tooth disease type 2. Asymp, asymptomatic. ATCP, arthropathy, tendinous calcinosis, and progeroid syndrome. CD, cardiac disease. CMD, congenital muscular dystrophy. DCM, dilated cardiomyopathy. EDMD, Emery-Dreifuss muscular dystrophy. FPLD2, familial partial lipodystrophy type 2. IRS, insulin resistance syndrome. LGMD, limb-girdle muscular dystrophy. MADA, mandibuloacral dysplasia type A. (C) Schematic depicting the frequency of variant alleles (denoted by short vertical lines). Alleles are grouped on the same horizontal line, based on whether they were identified in a single individual (‘1x’), two individuals (‘2x’) and so on. Most variants were rare (∼1–5 per 100,000 sequenced chromosomes) in the ExAC population. Relatively frequent variants included p.G638R (‘17x’), p.G602S (‘29x’) and p.R644H (‘47x’; ∼1–4 per 10,000 sequenced chromosomes) and p.R644C (‘145x’; >1 per 1,000 sequenced chromosomes).
