FIGURE 5.
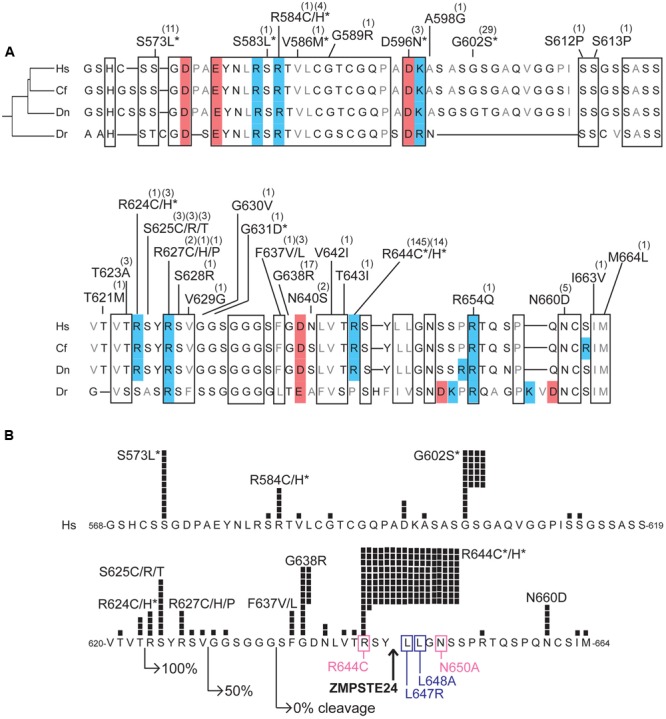
Variants located in the unstructured C-terminal region unique to prelamin A. (A) Aligned amino acid sequences of the C-terminal region of prelamin A (human residues 568–664) in Homo sapiens (‘Hs’; UniProt P02545), Camelus ferus (‘Cf’; camel; Refseq XP_014422471.1), Dasypus novemcinctus (‘Dn’; nine-banded armadillo; Refseq XP_004462616.1), and Danio rerio (‘Dr’; zebrafish; GenBank AAI63807.1). These animals were selected to highlight differences among LMNA residues in a diverse array of species; residues conserved among these species, aligned manually and using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) are likely to be functionally important. Highly conserved regions are boxed. Acidic residues are shaded red; basic residues are shaded blue. ExAC variants are indicated; parentheses show the number of affected individuals. Asterisks indicate variants also reported in the laminopathy database. (B) ExAC variants and number of affected individuals (black squares) in prelamin A residues 568–664, in the context of regions and residues important for ZMPSTE24-dependent cleavage. Right-pointing arrows indicate the N-terminal residue of fragments tested for ZMPSTE24-dependent cleavage in mammalian cells; GFP-fused fragments comprising residues 624–664 were cleaved efficiently (100%), residues 630–664 were cleaved inefficiently (50% cleaved) and residues 636-664 remained uncleaved (Barrowman et al., 2012). Upward arrow: ZMPSTE24 cleavage site. Missense mutations that reduce cleavage efficiency (pink box) or abrogated cleavage (purple box; Barrowman et al., 2012) are indicated. Asterisks indicate variants also reported in the laminopathy database.
