Abstract
Congenital adrenal hyperplasia, one of the most frequent autosome recessive disorders, is caused by defects in steroidogenic enzymes involved in the cortisol biosynthesis. Approximately 95% of the cases are caused by abnormal function of the 21-hydroxylase enzyme. This deficiency leads to androgen excess, consequently, to virilization and rapid somatic growth with accelerated skeletal maturation. Mutations in CYP21A2 are responsible for different forms of 21-hydroxylase deficiency. Mild impairment in the enzymatic activity causes the non-classic or late-onset congenital adrenal hyperplasia that is observed with a prevalence of 1 in 1000 subjects in different populations. The present paper describes a de novo mutation that occurred in the paternal meiosis. The child, who was conceived by in vitro fertilization, presented with precocious puberty and diagnosed with non-classical 21-hydroxylase deficiency. DNA sequencing showed the compound heterozygosis for a de novo CYP21A1P/A2 chimeric gene and the p.Val281Leu mutation inherited from her mother, who was heterozygous for the mutation. The chimeric gene showed pseudogene-derived sequence from 5′-end to intron 3 and CYP21A2 sequences from intron 3 to 3′-end of the gene. Sequencing analysis of the father did not show any mutation. The multiplex ligation-dependent probe amplification (MLPA) assay did not indicate loss of DNA discarding gene deletion but confirmed the chimeric gene. In addition, supernumerary copies of CYP21A1P were observed for both parents and for the affect child. Since paternity has been confirmed, those results suggest that a de novo large gene conversion in the paternal meiosis could have occurred by misalignment of alleles bearing different copy numbers of genes in CYP21 locus.
Keywords: CYP21A2 gene, CYP21A1 pseudo gene, 21-Hydroxylase deficiency, Congenital adrenal hyperplasia, In vitro fertilization, Non-classical CAH form
1. Introduction
Congenital Adrenal Hyperplasia (CAH), one of the most frequent autosome recessive disorders, is caused by defects in any of the steroidogenic enzymes involved in cortisol biosynthesis. The adrenal gland synthesizes three main classes of hormones: mineralocorticoids, glucocorticoids, and sex steroids. Other enzymes participate in several steps in adrenal steroids synthesis: steroidogenic acute regulatory protein (STAR), 3β-hydroxysteroid dehydrogenase (HSD3B2), 21-hydroxylase (CYP21A2), 11β-hydroxylase (CYP11B1) and 17α-hydroxilase (CYP17A1) [1]. Approximately 95% of CAH cases are caused by the deficiency of CYP21A2 [2].
As result of these enzyme deficiencies low or null production of cortisol and/or aldosterone is observed. Cortisol controls the hypothalamic-pituitary axis through a negative feedback mechanism. The impairment in cortisol production results in increased secretion of corticotrophin releasing hormone (CRH) and adrenocorticotropic hormone (ACTH) by the hypothalamus and the pituitary, respectively, causing super stimulation of adrenal glands and, consequently, their hyperplasia [3]. Depending on the affected enzyme, different forms of CAH manifest with distinct clinical symptoms. In the most frequent form, the CYP21A2 deficiency, aldosterone and cortisol pathways are blocked or partially impaired whereas the androgen pathway, which does not involve 21-hydroxylation, is overstimulated and consequently the synthesis of androgen increases [1], [4].
21-Hydroxylase deficiency (OMIM #201910) is divided in classical and non-classical forms depending on the severity of the clinical features and vary according to the level of residual activity of the enzyme. The classical form presents at birth and is characterized by total or almost total impairment of the enzymatic activity. Usually, it leads to androgen excess causing progressive virilization with or without concomitant salt loss and is classified as salt-wasting or simple virilizing form, respectively. The non-classical form, in which more than 10% of CYP21A2 enzymatic activity is retained, is generally less severe than the classical form, with late phenotypic manifestation such as precocious puberty and some secondary characteristics as hirsurtism and menstrual disorders in females or it can remain asymptomatic [1], [5], [6], [7].
General data have indicated that the incidence of the classical form is 1:10,000 to 1:15,000 live births, whereas the non-classical form shows a higher prevalence, occurring with an estimated incidence of 1:1000 births in Caucasian populations [2], [8], [9].
The CYP21A2 gene is located within the HLA major histocompatibility complex class III (MHC class III), mapping to the short arm of chromosome 6 (6p21.3), within a 2.7 kb chromosomal region divided in 10 exons [10]. In human MHC class III region, the following genes are arranged in tandem: serine/threonine kinase (RP), complement C4, steroid 21-hydroxylase (CYP21) and tenascin (TNX); they form a genetic unit designated RCCX module [11], [12]. Alleles bearing one copy of each CYP21A2 active form and CYP21A1P nonfunctional pseudogene, are bimodular, considered the most frequent arrangement. However, monomodular, trimodular and tetramodular normal alleles have been observed [4], [10], [11]. Although CYP21A2 and CYP21A1P have approximately 98% of homology in exons and 96% in introns, CYP21A1P is nonfunctional because it bears several deleterious mutations [13], [14].
Mutations in CYP21A2 are responsible for different forms of CYP21A2 deficiency, depending on how they affect the protein structure, the enzyme production or the enzymatic activity. One consequence of CYP21 and C4 genes arranged in tandem is to facilitate unequal crossing-over events in this region [4], [15]. Therefore, misaligned units may recombine during meiosis and lead to the formation of gametes with different number of gene units [16]. Therefore, the first molecular defect associated with CAH was a 30-kb deletion forming an allele that carried an inactive CYP21A1P/A2 chimeric gene [17], [18]. The second molecular defect described was also result of unequal recombination; known as large gene conversion it also resulted in an inactive CYP21A1P/A2 chimeric gene, but without loosing DNA [19], [20], [21], [22]. Those two mutations were identified in only 8–12% of cases of CYP21A2 deficiency in Brazil [23], [24], [25]. CYP21A1P/A2 chimeric genes with different sequence compositions have been described depending on the recombination breakpoint [22], [26].
In this paper we describe a de novo large gene conversion identified in a patient conceived by in vitro fertilization (IVF). This mutated allele might have formed in the paternal meiosis as a result of sequence misalignment in alleles containing different number of gene copies within the CYP21 locus.
2. Materials and methods
2.1. Subject
The patient was a daughter of non-consanguineous parents that had been conceived by in vitro fertilization (IVF). She was referred for investigation because premature pubarche at the age of eight years and seven months, with a history of premature pubic hair development and axillary odor since seven years and ten months old, with rapid evolution. Thelarche was denied. At physical examination, her height and weight were 130.3 cm (P50) and 32.3 kg (P85), respectively, and Tanner stage was P3/M1. Table 1 shows laboratorial and radiological investigation.
Table 1.
Hormonal data and bone age presented by the affected child and reference values.
| Reference value | Patient | |
|---|---|---|
| 17-Hydroxyprogesterone (17OHP) (ng/mL) | 0.1–1.39 | 20.7 |
| Testosterone (ng/dL) | < 20.0 | 58.6 |
| SDHEA (μg/dL) | 8.0–130.0 | 94.3 |
| Androstenedione (ng/mL) | < 1.6 | 2 |
| Bone age years | 8.6 | 10 |
The background to the family history shows that her father has two healthy children from a previous marriage.
2.2. Molecular analysis
Genomic DNAs of the patient and her parents were obtained from peripheral blood samples by Proteinase K digestion and phenol/chloroform extraction following standard techniques [27]. CYP21A2 was specifically amplified in two or four fragments, depending on the presence or absence of intron 2 variant c.290-13A > C, using specific CYP21A2 primers (Table 2). The amplified fragments were gel purified with Wizard SV Gel and PCR Clean-UP System (Promega®) and directly sequenced with Big Dye Terminator Cycle Sequencing Kit V3.1 Ready Reaction (ABI PRISM/PE Biosystems, Foster City, CA, USA) using internal primers in addition to sense and antisense primers used in the allele-specific PCRs (Table 2). Purified PCR products were sequenced in both sense and antisense orientations in an ABI PRISM 3100 Automated DNA Sequencer according to the manufacturer's recommendations (ABI PRISM/PE Biosystems, Foster City, CA, USA). Nucleotide and amino acid numbering followed the published sequence by Higashi et al. [13] (NCBI #M12792).
Table 2.
Primers used in this study.
| Primer | Sequence (5′–3′) | Purpose |
|---|---|---|
| 5′21B1s_CYP21A2 | TGA GTG AGT GCC CAC AAA G | PCR, sequencing |
| ProEx1s_CYP21A2 | TTG AGT GAG TGC CCA CAA AGC | PCR, sequencing |
| Int1s_CYP21A2a | TGA GAG GCT GAT CTC GCT C | Sequencing |
| Int1as_CYP21A2a | CAG CCA AAG CAG CGT CAG CC | Sequencing |
| Int2Cs_CYP21A2 | CAC CCT CCA GCC CCC AC | PCR, sequencing |
| Int2Cas_CYP21A2 | CAG CTT GTC TGC AGG AGG AGG | PCR, sequencing |
| Ex2as_CYP21A2a | TAA GTG GCT CAG GTC TGC C | Sequencing |
| Ex3ns_CYP21A2 | ACC TGT CCT TGG GAG ACT AC | PCR, sequencing |
| Ex6nas_CYP21A2 | CCT CAG CTG CAT CTC CAC GA | PCR, sequencing |
| Int7as_CYP21A2a | CAG AGC TGA GTG AGG GTG | Sequencing |
| Ex9s_CYP21A2a | TTG GGG ATG AGT GAG GAA AG | Sequencing |
| Ex9as_CYP21A2a | CTG GAG TTC TTG CCT GGC TC | Sequencing |
| Ex10s_CYP21A2a | GGA GCT CTT CGT GGT GCT GA | Sequencing |
| Int10as_CYP21A2 | CAG AGG GAG CTG GAG TTG A | PCR, sequencing |
Internal primers.
2.3. Multiplex ligation-dependent probe amplification analysis (MLPA)
DNA from the patient and her parents was also examined by the multiplex ligation-dependent probe amplification (MLPA) technique using the SALSA MLPA probemix P050-C1 CAH (MRC Holland, Amsterdam, Netherlands®). The kit contains 27 MLPA probes and 10 control fragments. Eight probes are specific for CYP21A2 and 4 probes are specific for CYP21A1P. CYP21A2 probes detect the following mutations: 8-bp deletion in exon 3, p.Ile172Asn in exon 4, p.Val237Glu and p.Met239Lys in exon 6 and c.920_921insT in exon 7; in addition, it is provided probes for c.290-13A > C and c.-113C > T SNP in intron 2 and 5′ region, respectively. Furthermore, this kit contains 6 probes for TNXB gene and 1 probe for ATF6B in order to define CYP21A2 gene deletions. Eight references probes detecting sequences on different chromosomes are provided for data internal normalization. An Excel-based spreadsheet constructed for the analysis was used. Data have been normalized using five bimodular controls.
2.4. Ethical
This study was approved by the Ethics Committee from Universidade Estadual de Campinas (São Paulo, Brasil) and Universidade Federal do Triângulo Mineiro (Uberaba, Minas Gerais, Brasil) and an informed consent was obtained from the relatives.
3. Results
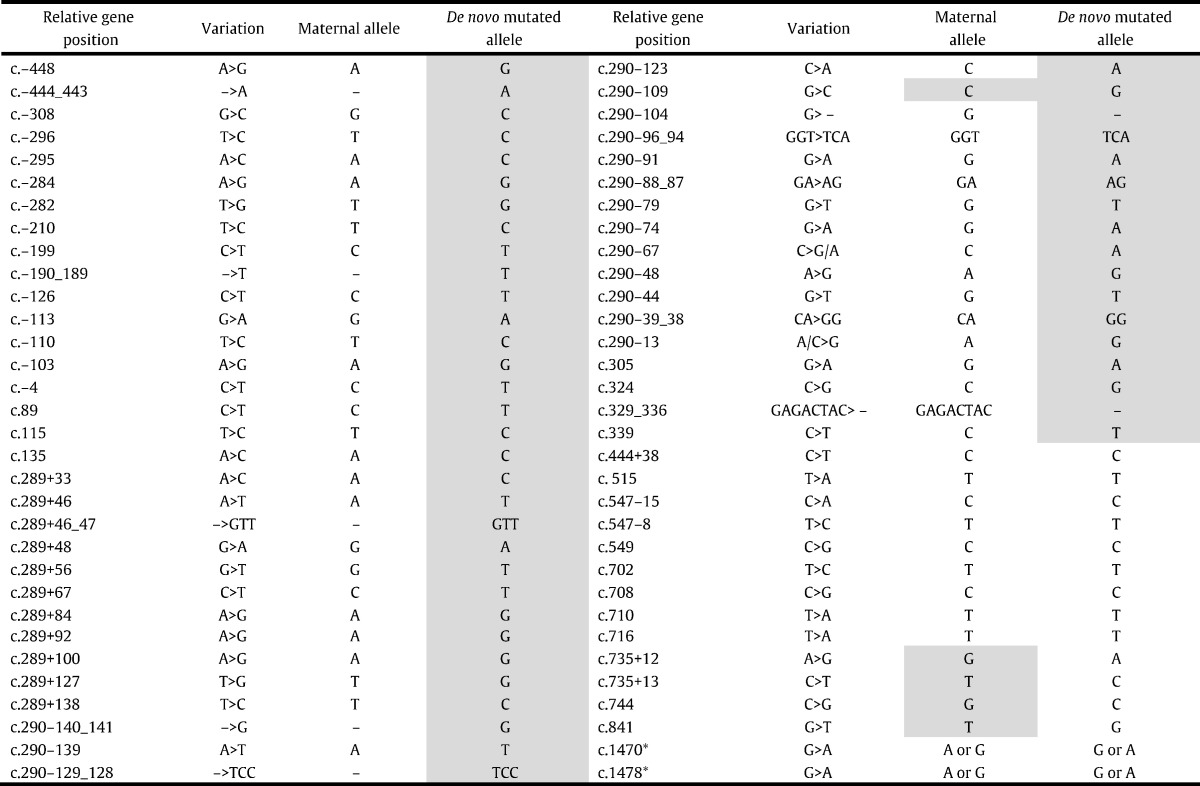
Direct sequencing analyses of CYP21A2 gene of the patient identified heterozygosity for p.Val281Leu mutation and for all pseudogene-derived sequences from c.-620 to intron 3 including c.290-13A/C > G (intron 2 Splice) and c.329_336delGAGACTAC (exon 3 delta 8) mutations suggesting a large gene conversion or a 30-kb deletion with CYP21A1P/A2 chimeric gene (Table 3). The mutation p.Val281Leu was also identified in the mother, however no deleterious mutations had been found in the father's gene. Paternity had been confirmed with different VNTR (data not shown).
Table 3.
CYP21A2 sequencing data for the patient.
Gray shading corresponds to pseudogene-derived sequences, except c.290–109 that has not been annotated in the pseudogene but is a p.Val281Leu marker; c.841 corresponds to p.Val281Leu mutation. *Father and mother are also heterozygous for those polymorphic sequence variations, therefore segregation has not been determined.
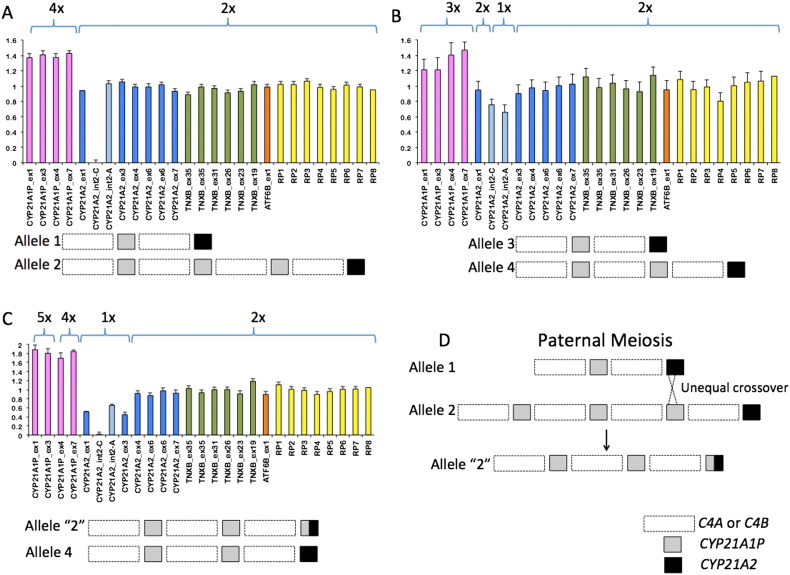
MLPA confirmed a possible large gene conversion generating a CYP21A1P/A2 gene since it showed diminished hybridization signals with CYP21A2 probes from 5′ to exon 3, but the hybridization signals of the corresponding pseudogene probes were visibly increased (Fig. 1C). Conversely, neither the father nor the mother MLPA results showed diminished hybridization signals for CYP21A2 probes (Fig. 1A, B). Those results also demonstrated increased signals for CYP21A1P probes for both parents and the child indicating that they carry alleles with supernumerary pseudogene copies.
Fig. 1.
Integrated and normalized MLPA data and a proposal for the formation of a de novo CYP21A1P/A2 chimeric gene. MLPA graphics obtained for the father (A), the mother (B) and the patient (C) are shown; columns correspond to integrated and normalized electropherogram peak areas for each probe indicated below; values of 1.0 ± 0.2 indicate two copies; deletions (1 copy) or duplications (2 or more copies) are indicated by intensities < 0.5 or > 1.2, respectively. Pink = CYP21A1P probes; blue = CYP21A2 probes; green = TNXB probes; orange = ATF6B probe; yellow = control probes on different chromosomes. Schemes below each graphic illustrate the composition of each allele in the family regarding copy number of C4 and CYP genes. In (D) is represented the probable rearrangement occurred in the paternal meiosis to generate an allele bearing the CYP21A1P/A2 chimeric gene.
4. Discussion
Most CYP21A2 deficient alleles carry pre-existing mutations in the homolog pseudogene, located in tandem alternated with C4A and C4B genes. In general, mutant alleles are inherited from carrier parents. Novel or de novo mutations acquired during gametogenesis or fetal development are less observed, but can occur in rates of ~ 5% and ~ 2%, respectively [28].
Individuals with non-classical CYP21A2 deficiency are frequently homozygous for p.Val281Leu mutation or compound heterozygous bearing different CYP21A2 mutations on each allele [29]. The missense mutation p.Val281Leu accounts for at least one of the CYP21A2 alleles in most non-classical genotypes. The patient we described here inherited the p.Val281Leu from her mother whereas the other allele bears a chimeric CYP21A1P/A2 gene with pseudogene sequences from the 5′ end to intron 3 and CYP21A2 sequences from intron 3 to the 3′ end of the gene. Since this chimeric gene is absent in both parents' genotypes we inferred that it arose as a de novo mutation.
MLPA data showed supernumerary copies of pseudogene in both father and mother genotypes. It is well known that such alleles favor unequal crossovers during meiosis generating chimeric genes but in general those rearrangements are not frequently observed [30]. CYP21A1P duplication associated to p.Val281Leu mutation is typical and corresponds to 98% of alleles bearing this mutation [31]. However, such duplication is not always associated with mutations in the CYP21A2 gene [32], as could be observed in father's results. The MLPA probe signal intensities for CYP21A1P in the affected child suggest four copies, two inherited from the mother and two inherited from the father indicating that the chimeric gene might be generated in association with two pseudogenes in the same allele. To illustrate the hypothesis of unequal crossover in the paternal gametogenesis, we propose a genotype with a bimodular and a tetramodular allele for the father as shown in Fig. 1D. A bimodular gamete with CYP21A1P/A2 chimeric gene can be generated if unequal crossover occurs between CYP21A2 in allele 1 and CYP21A1P in allele 2.
In sperm, spontaneous recombination between of CYP21A2 and CYP21A1P (i.e., gene conversions or deletions) are detected in 1:1000 to 1:100,000 cells [33]. In patients, 1 to 2% of affected alleles result from spontaneous mutations when neither parent is a carrier [34]. The gene conversions can occur during meiosis and mitosis, or perhaps only during mitosis [33]. It is well known that unequal crossing over facilitates large structural rearrangements resulting in copy number variations [35], whereas gene conversions promote relatively short sequence transfers [36].
Complex rearrangements involving the RCCX module had been observed. The heterozygosis for RCCX modules with different copy number variants has been considered to promote unequal crossovers during meiosis, leading to gene deletions, gene duplications and disease associations [12], [37].
Chung et al. [38] observed C4 genes organized in different configurations within RCCX modules when they studied complement C4 polymorphisms considering gene sizes, gene numbers and their orders. They identified haplotypes with one, two, three and four C4 genes, and the variability in the dosage of CYP21A1P pseudogene and CYP21A2 gene. They concluded that quadrimodular structures could have been generated by unequal crossover between a trimodular and a regular bimodular partner with one CYP21A1P and one CYP21A2. Similarly, we propose that an unequal crossover generated a de novo chimeric gene identified in the affected child.
5. Conclusion
The genotype presented by the patient is consistent with the non-classic 21-OH deficiency. However, the CYP21A1P/A2 chimeric gene identified in the affect child was not present in either parent's genotypes. To our knowledge it is the first report on a case of in vitro conceived child bearing a de novo CYP21A2 mutation. Therefore, this study illustrates the importance of the molecular analyses not only for the affected child but also for the parents, especially for guiding pre-natal diagnosis and genetic counseling. Since alleles with pseudogene supernumerary copies is present in all populations we recommend that the risk of unequal crossover generating chimeric genes is considered for genetic counseling purposes. Despite all the modern techniques available, it is still impossible to predict if a genetic recombination will occur in germ cells. However, due to the increased risk observed with supernumerary gene copy alleles it would be advisable to offer CYP21A2 genotyping whether for couples with spontaneous gestation or those submitted to in vitro fertilization.
Conflict of interests
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Acknowledgments
The authors want to acknowledge Cristiane dos Santos Cruz Piveta for the technical support. This study was funded by FAPESP grants #s 2011/5180-2 to MPdeM and FAPESP Doctoral Fellowship # 2012/16815-0 to DPM.
References
- 1.Nimkarn S., New M.I. 21-Hydroxylase-deficient congenital adrenal hyperplasia. Gene Reviews®[Internet] 2013 (Last Update: August 29) [Google Scholar]
- 2.Nimkarn S., Lin-Su K., New M.I. Steroid 21-hydroxylase deficiency congenital adrenal hyperplasia. Pediatr. Clin. N. Am. 2011;58:1281–1300. doi: 10.1016/j.pcl.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 3.White P.C., Speiser P.W. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr. Rev. 2000;21:245–291. doi: 10.1210/edrv.21.3.0398. [DOI] [PubMed] [Google Scholar]
- 4.Kamrath C., Hartmann M.F., Wudy S.A. Androgen synthesis in patients with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Horm. Metab. Res. 2013;45:86–91. doi: 10.1055/s-0032-1331751. [DOI] [PubMed] [Google Scholar]
- 5.Miller W.L. Congenital adrenal hyperplasias. Endocrinol. Metab. Clin. N. Am. 1991;20:721–740. [PubMed] [Google Scholar]
- 6.Lin-Su K., Nimkarn S., New M.I. Congenital adrenal hyperplasia in adolescents: diagnosis and management. Ann. N. Y. Acad. Sci. 2008;1135:95–98. doi: 10.1196/annals.1429.021. [DOI] [PubMed] [Google Scholar]
- 7.Witchel S.F. Non-classic congenital adrenal hyperplasia. Steroids. 2013;78:747–750. doi: 10.1016/j.steroids.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Trakakis E., Basios G., Trompoukis P., Labos G., Grammatikakis I., Kassanos D. An update to 21-hydroxylase deficient congenital adrenal hyperplasia. Gynecol. Endocrinol. 2009;3:1–9. doi: 10.3109/09513590903015494. [DOI] [PubMed] [Google Scholar]
- 9.Wedell A. Molecular genetics of 21-hydroxylase deficiency. Endocr. Dev. 2011;20:80–87. doi: 10.1159/000321223. [DOI] [PubMed] [Google Scholar]
- 10.White P.C., Grossberger D., Onufer B.J. Two genes encoding steroid 21-hydroxylase are located near the genes encoding the fourth component of complement in man. Proc. Natl. Acad. Sci. U. S. A. 1985;82:1089–1093. doi: 10.1073/pnas.82.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haglund-Stengler B., Martin Ritzen E., Gustafsson J., Luthman H. Haplotypes of the steroid 21-hydroxylase gene region encoding mild steroid 21-hydroxylase deficiency. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8352–8356. doi: 10.1073/pnas.88.19.8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z., Mendoza A.R., Welch T.R., Zipf W.B., Yu C.Y. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J. Biol. Chem. 1999;274:12147–12156. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- 13.Higashi Y., Yoshioka H., Yamane M., Gotoh O., Fuji-Kuriyama Y. Complete nucleotide sequence of two steroid 21-hydroxylase genes tandemly arranged in human chromosome: a pseudogene and genuine gene. Proc. Natl. Acad. Sci. U. S. A. 1986;83:2841–2845. doi: 10.1073/pnas.83.9.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White P.C., New M.I., Doupont B. Structure of human steroid 21-hydroxylase genes. Proc. Natl. Acad. Sci. U. S. A. 1986;83:5111–5115. doi: 10.1073/pnas.83.14.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strachan T., White P.C. Molecular pathology of steroid 21-hydroxylase deficiency. J. Steroid Biochem. Mol. Biol. 1991;40:537–543. doi: 10.1016/0960-0760(91)90274-9. [DOI] [PubMed] [Google Scholar]
- 16.Werkmeister J.W., New M.I., Dupont B., White P.C. Frequent deletion and duplication of the steroid 21-hydroxylase genes. Am. J. Hum. Genet. 1986;39:461–469. [PMC free article] [PubMed] [Google Scholar]
- 17.Carrol M.C., Campbell R.D., Porter R.P. Mapping of steroid 21-hydroxilase genes adjacent to complement component C4 genes in HLA, the major histocompatibility complex in man. Proc. Natl. Acad. Sci. U. S. A. 1985;82:521–525. doi: 10.1073/pnas.82.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rumsby G., Carrol M.C., Porter R.R., Grant D.B., Hjelm M. Deletion of the steroid 21-hydroxylase and complement C4 genes in congenital adrenal hyperplasia. J. Med. Genet. 1986;23:204–209. doi: 10.1136/jmg.23.3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada F., Kimura A., Iwanaga T., Shimozawa K., Yata J., Sasazuki T. Gene conversion-like events cause steroid 21-hydroxylase deficiency in congenital adrenal hyperplasia. Proc. Natl. Acad. Sci. U. S. A. 1987;84:8091–8094. doi: 10.1073/pnas.84.22.8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urabe K., Kimura A., Harada F., Iwanaga T., Sasazuki T. Gene conversion in steroid 21-hydroxylase genes. Am. J. Hum. Genet. 1990;46:1178–1186. [PMC free article] [PubMed] [Google Scholar]
- 21.Collier S., Tassabehji M., Sinnott P., Strachan T. A de novo pathological point mutation at the 21-hydroxylase locus: implications for gene conversion in the human genome. Nat. Genet. 1993;3:260–265. doi: 10.1038/ng0393-260. [DOI] [PubMed] [Google Scholar]
- 22.Concolino P., Mello E., Minucci A., Giardina E., Zuppi C., Toscano V., Capoluongo E. A new CYP21A2/CYP21A1P chimeric gene identified in an Italian woman suffering from classical congenital adrenal hyperplasia form. BMC Med. Genet. 2009;10:1–7. doi: 10.1186/1471-2350-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De-Araujo M., Sanches M.R., Suzuki L.A., Guerra G., Jr., Farah S.B., De-Mello M.P. Molecular analysis of CYP21 and C4 genes in Brazilian families with the classical form of steroid 21-hydroxylase deficiency. Braz. J. Med. Biol. Res. 1996;29:1–13. [PubMed] [Google Scholar]
- 24.Bachega T.A., Billerbeck A.E., Madureira G. Molecular genotyping in Brazilian patients with the classical and nonclassical forms of 21-hydroxylase deficiency. J. Clin. Endocrinol. Metab. 1998;83:4416–4419. doi: 10.1210/jcem.83.12.5350. [DOI] [PubMed] [Google Scholar]
- 25.Paulino L.C., Araujo M., Guerra G., Jr., Marini S.H., De Mello M.P. Mutation distribution and CYP21/C4 locus variability in Brazilian families with the classical form of the 21-hydroxylase deficiency. Acta Paediatr. 1999;88:275–283. doi: 10.1080/08035259950170024. [DOI] [PubMed] [Google Scholar]
- 26.Lee H.-H. Chimeric CYP21P/CYP21 and TNXA/TNXB genes in the RCCX module. Mol. Genet. Metab. 2005;84:4–8. doi: 10.1016/j.ymgme.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J., Fritsch E.F., Maniatis T.E. Cold Spring Harbor; New York: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 28.Levo A., Partanen J. Novel mutations in the human CYP21 gene. Prenat. Diagn. 2001;21:885–889. doi: 10.1002/pd.167. [DOI] [PubMed] [Google Scholar]
- 29.Witchel S.F., Azziz R. Nonclassic congenital adrenal hyperplasia. Int. J. Pediatr. Endocrinol. 2010;2010:625105. doi: 10.1155/2010/625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumgartner-Parzer S.M., Fischer G., Vierhapper H. Predisposition for de novo gene aberrations in the offspring of mothers with a duplicated CYP21A2 gene. J. Clin. Endocrinol. Metab. 2007;92:1164–1167. doi: 10.1210/jc.2006-2189. [DOI] [PubMed] [Google Scholar]
- 31.Speiser P.W., New M.I., White P.C. Molecular genetic analysis of nonclassic steroid 21-hydroxylase deficiency associated with HLA-B14, DR1. N. Engl. J. Med. 1988;319:19–23. doi: 10.1056/NEJM198807073190104. [DOI] [PubMed] [Google Scholar]
- 32.Wedell A., Ritzen E.M., Haglund-Stengler B., Luthman H. Steroid 21-hydroxylase deficiency: three additional mutated alleles and establishment of phenotype genotype relationships of common mutations. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7232–7236. doi: 10.1073/pnas.89.15.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tusie-Luna M.T., White P.C. Gene conversions and unequal crossovers between CYP21 (steroid 21-hydroxylase gene) and CYP21P involve different mechanisms. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10796–10800. doi: 10.1073/pnas.92.23.10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speiser P.W., Dupont J., Zhu D. Disease expression and molecular genotype in congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J. Clin. Invest. 1992;90:584–595. doi: 10.1172/JCI115897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 36.Chen M.J., Cooper D.N., Chuzhanova N., Férec C., Patrinos G.P. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 37.Blanchong C.A., Zhou B., Rupert K.L. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in Caucasians: the load of RCCX genetic diversity on MHC-associated disease. J. Exp. Med. 2000;191:2183–2196. doi: 10.1084/jem.191.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung E.K., Yang Y., Rennebohm R.M. Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am. J. Hum. Genet. 2002;71:823–837. doi: 10.1086/342777. [DOI] [PMC free article] [PubMed] [Google Scholar]