Figure 3. A structure-based mechanism for SM protein-mediated SNARE complex assembly.
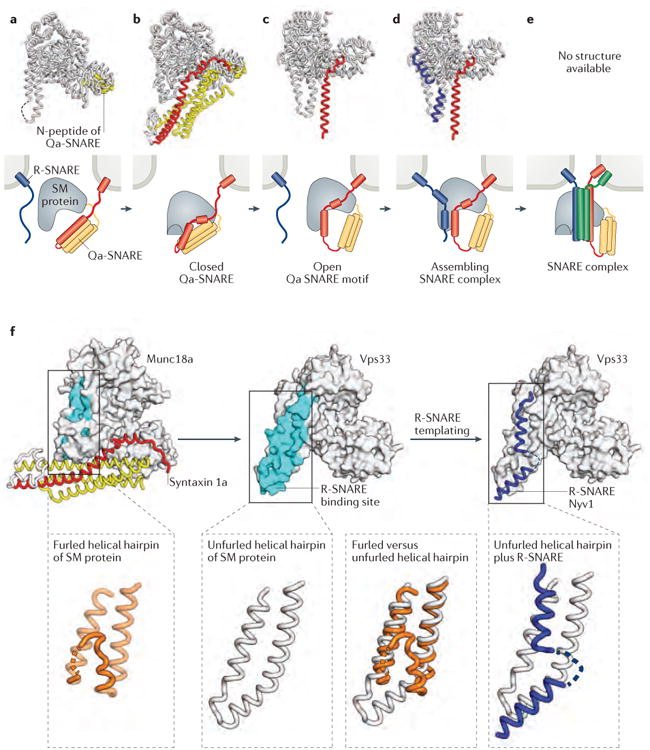
a-e | X-ray crystal structures (first row) and schematic representations (second row) of SM protein–SNARE complexes. The structures and schematics are placed in a speculative order to suggest a potential pathway for SM protein-templated SNARE complex assembly. a | Many SM proteins bind to the N-peptide lying at the extreme N-terminus of Qa- (or syntaxin-like) SNAREs. Shown is the structure of the SM protein Munc18a (grey) bound to the N-peptide of syntaxin 4 (yellow) (PDB code 3PUJ)92. b | Some Qa-SNAREs adopt an auto-inhibited conformation that binds to, and is stabilized by, the cognate SM protein. This binding mode requires a furled conformation for the helical hairpin of the SM protein. Shown is the structure of Munc18a bound to the cytoplasmic portion of syntaxin 1a (yellow and red) (PDB code 3C98)94. c | The opening of the Qa-SNARE is accompanied by the unfurling of the SM protein's helical hairpin, which exposes the R-SNARE binding site. Shown is the structure of the SM protein Vps33 (grey) bound to the SNARE motif of the Qa-SNARE Vam3 (red) (PDB code 5BUZ)12. d | Binding of the R-SNARE to the SM protein leads to a half-zippered SNARE complex representing a potential early intermediate in SNARE complex assembly. Shown is a model that combines the structures of Vps33 bound to the R-SNARE Nyv1 (blue) (PDB code 5BV0) and Vp33 bound to the Qa-SNARE Vam3 (red and yellow) (PDB code 5BUZ)12. e | After formation of the trans-SNARE complex, SM proteins may bind directly to the resulting four-helix bundle. The resulting complex has not, however, been structurally defined. f | Unfurling of the domain 3a helical hairpin of the SM protein reveals the R-SNARE binding site.
