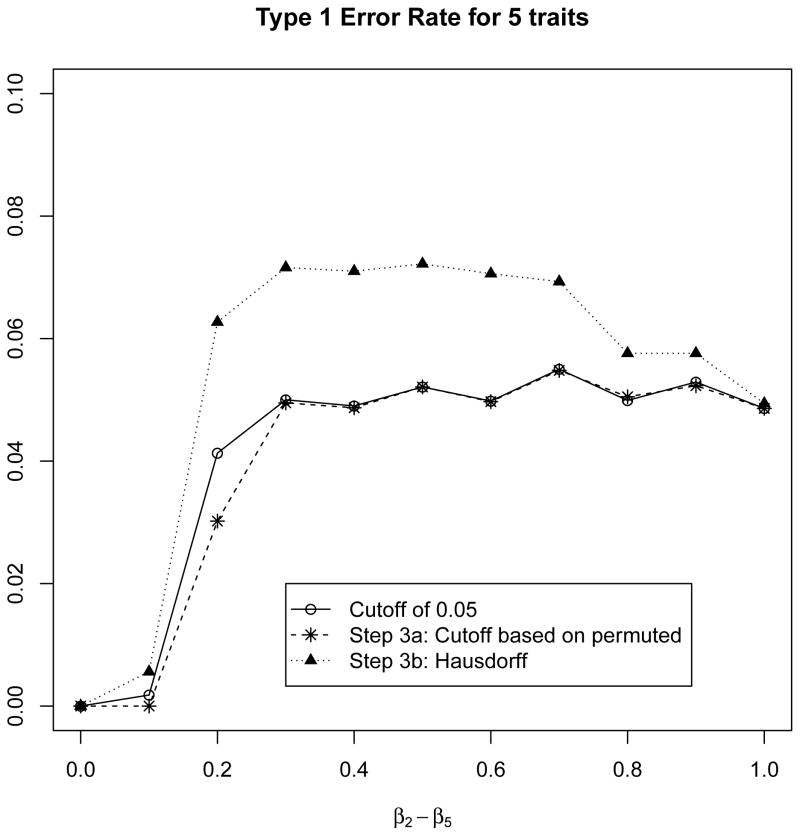
Figure 2.
For 5 normally distributed phenotypes, we evaluated the type-1 error rate of the proposed approaches when one phenotype is not associated with the SNP, but the other 4 phenotypes are strongly associated with the SNP. We fixed β1 = 0 and β2 = β3 = β4 = β5 vary from 0 to 1 by 0.1. Note that the ad hoc approach and the cut-off based permutation approaches both maintain the type 1 error rate, but the Hausdorff based approach has a slightly inflated type 1 error rate of 0.06 to 0.07.