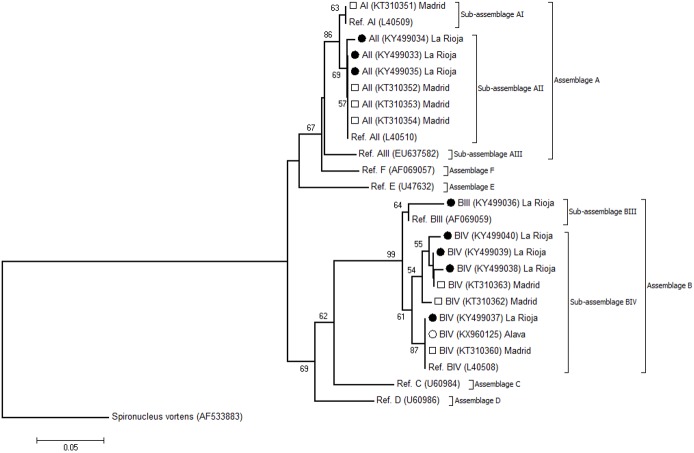
Fig 3. Evolutionary relationships among G. duodenalis sub-assemblages at the GDH locus inferred by a Neighbor-Joining analysis of the nucleotide sequence covering a 383-bp region (positions 88 to 470 of GenBank accession number L40508) of the gene.
The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 iterations) is indicated next to the branches. Bootstrap values lower than 50% were not displayed. The evolutionary distances were computed using the Kimura 2-parameter method. The rate variation among sites was modelled with a gamma distribution (shape parameter = 2). Filled circles represent AII, BIII, and BIV sequences generated in this study. Open circles indicate G. duodenalis sequences of human origin previously reported in other Spanish regions that were included in the analysis for comparative purposes. Spironucleus vortens was used as outgroup taxa.