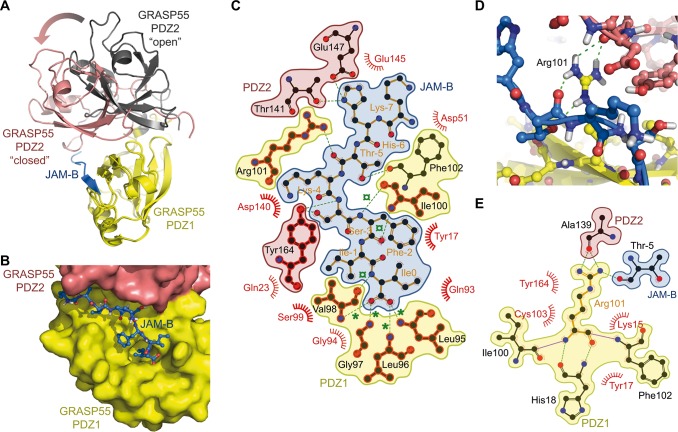
Fig 5. Co-Crystal structure of GRASP55 with JAM peptides.
(A) Superimposition of the previously reported ‘ligand-free’ GRASP55 PDZ12 X-ray structure (PDB ID: 3RLE) with ‘cargo-bound’ complex, showing GRASP55 PDZ12 structure bound to JAM-B (PDB ID: 5GMI). The ‘ligand free’ published GRASP55 PDZ12 structure is displayed as yellow (PDZ1) and black (PDZ2) ribbons, while the ‘cargo-bound’ conformation is shown as yellow (PDZ1) and salmon (PDZ2); the JAM-B peptide is indicated in blue. The arrow highlights the 33-degree global rotation of the PDZ2 domain after aligning both PDZ1 domains. (B) Surface representation of the GRASP55 PDZ12/JAM-B complex structure showing the JAM-B peptide embedded in the groove formed by the GRASP55 PDZ1 (yellow) and PDZ2 (salmon) domains. (C) LigPlot+ representation of the complex pinpointing the hydrogen bonding interaction contacts (green) as well as the van der Waals and hydrophobic contacts (semicircles). Green stars (*) highlight carboxylic hydrogen bond contacts in the binding pocket, and green cross circles (¤) indicate β-sheet hydrogen bonding interactions. Conserved contact residues in the JAM-B and JAM-C complex structures are highlighted in red. (D) Detailed view of the Arg101 hydrogen bonding interaction network with the PDZ2 domain (salmon) and JAM-B (blue), as revealed by the X-rays structure. (E) LigPlot+ representation of the PDZ1 Arg101 hydrogen bonding network with JAM-B and PDZ2. The color coding is the same as that described in (C).