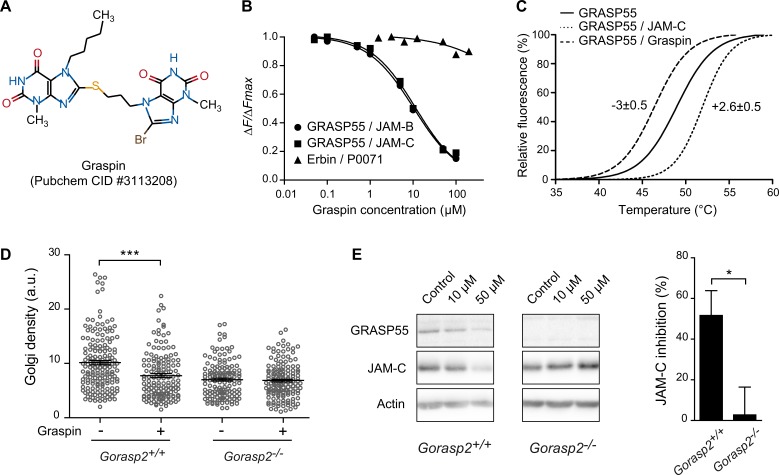
Fig 6. Characterization of a small compound inhibiting GRASP55 interaction with JAMs.
(A) 2D structure, PubChem Common Identifier (CID) and given name of the prioritized hit from the structure-based drug design approach. (B) Representative curves were obtained by HTRF using GST-GRASP55 FL or GST-Erbin with the indicated biotinylated peptides and competed with Graspin (IC50 = 8.4 μM for GRASP55/JAM-B and 12 μM for GRASP55/JAM-C). (C) Fluorescence profiles presenting results obtained by differential scanning fluorimetry (DSF). Shifts in the melting temperatures of His-GRASP55 PDZ12 incubated with a twelve molar equivalent of ligand or DMSO as control are shown. (D) Golgi density of Gorasp2+/+ and Gorasp2-/- MEFs treated or not with Graspin at 50 μM during 48 h. Each circle represents one Golgi. Data are the mean ± s.e.m. of pooled results of three independent experiments (analysis of 30–90 Golgi per condition, per experiment). Student’s unpaired t-test; ***: P<0.001. (E) Left panel, immunoblot for GRASP55 and JAM-C following exposure of Gorasp2+/+ and Gorasp2-/- MEFs to increasing doses of Graspin. Right panel, quantification of JAM-C expression in Gorasp2+/+ and Gorasp2-/- MEFs treated with Graspin at 50 μM for 48 h. Data are expressed as mean of JAM-C inhibition as compared to control condition (vehicle) in five independent experiments. Student’s unpaired t-test; *: P<0.05.