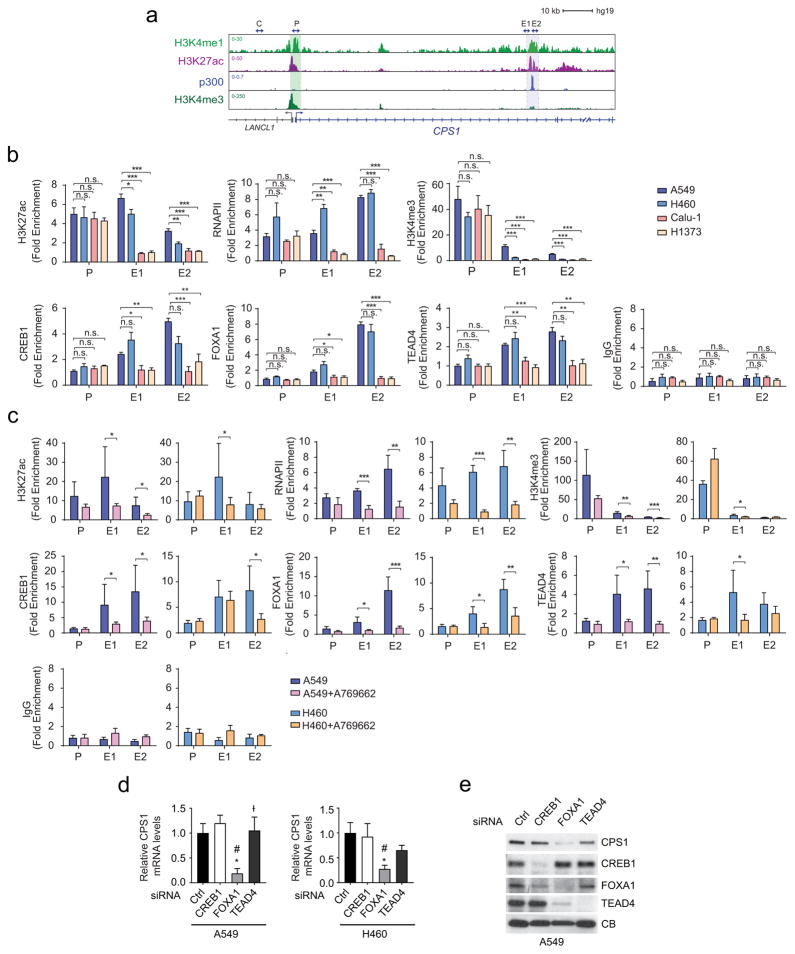
Extended Data Fig. 6. LKB1 regulates CPS1 transcription via AMPK-mediated effects.
a, Chromatin signatures at the CPS1 locus in A549 cells. Promoter and enhancer sequences are shaded. Arrowheads indicate amplicons for control (C), promoter (P) and enhancer (E1, E2) regions for ChIP-qPCR in b and c. b, Chromatin occupancy of H3K27ac, RNAPII, H3K4me3, CREB1, FOXA1, TEAD4 and IgG (negative control) in KL (A459, H460) and in K (Calu-1, H1373) cells. Data are the average and SD of two independent cultures, each with two technical replicates (total n=4). c, Chromatin occupancy of H3K27ac, RNAPII, H3K4me3, CREB1, FOXA1, TEAD4 and IgG in A549 and H460 cells treated with DMSO or 250μM A769662. Data are the average and SD of three independent cultures, each with two technical replicates (total n=6). d, Effects of CREB1, FOXA1, and TEAD4 silencing on CPS1 mRNA expression in A549 and H460 cells. Data are the average and SD of three or more replicates. e, Abundance of CPS1, CREB1, FOXA1, and TEAD4 in A549 cells. CB was used as a loading control. In b and c, statistical significance was assessed using two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p< 0.001, n.s. not significant. In d, statistical significance was assessed using one-way ANOVA followed by Tukey’s multiple comparisons test. *, p<0.05 comparing to control siRNA; #, p<0.05 comparing to siCREB1; Ɨ, p<0.05 comparing to siFOXA1. ChIP-qPCR in b was performed twice. All other experiments were repeated three times or more.