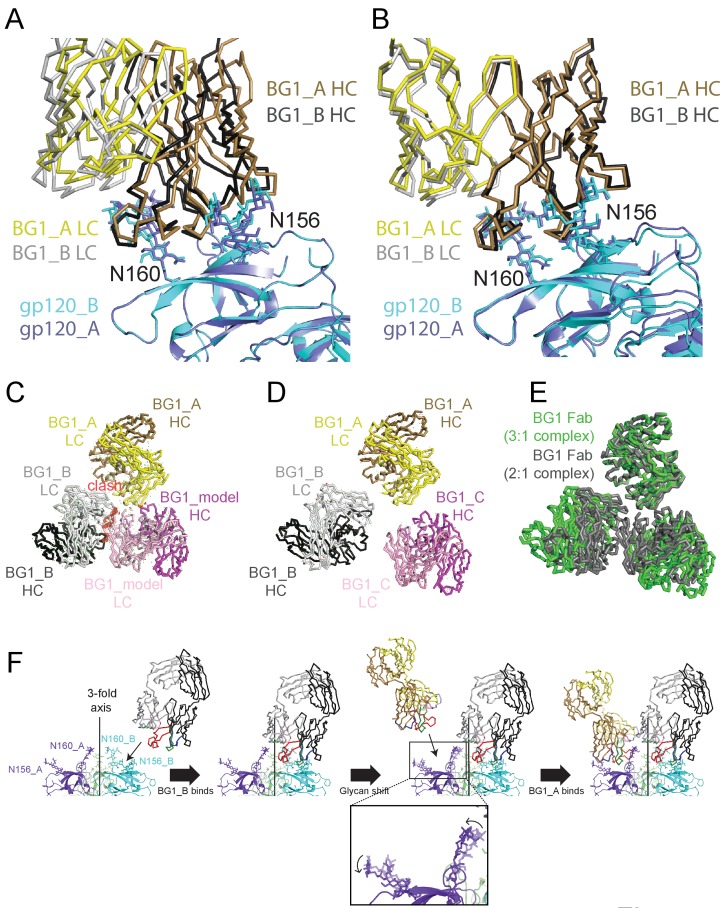
Figure 7. Asymmetry of BG1 interactions with Env trimer.
(A,B) Superposition of the two BG1 binding sites in the 2:1 BG1-Env structure by aligning either the V1V2 regions of gp120_A and gp120_B (panel A) or the Asn156gp120 and Asn160gp120 glycans within gp120_A and gp120_B (panel B). (C) Hypothetical model of three BG1 Fabs bound per Env trimer based on Fab orientations in the 2:1 BG1-Env structure. The BG1_A and BG_1 B Fabs are shown in their respective positions on the gp120_A and gp120_B subunits of Env trimer (view looking down the trimer three-fold axis). A third BG1 Fab (BG1 model) was positioned onto the gp120_C subunit assuming that it would interact with the Asn156gp120 and Asn160gp120 glycans in the common interaction observed for BG1_A and BG1_B (panel B). Predicted van der Waals clashes (red dots) were calculated using the show_bumps module in Pymol (Schrödinger, 2011). (D) Positions of three BG1 Fabs in the 27 Å 3:1 BG1-Env structure (no predicted van der Waals clashes). (E) Comparison of BG1 Fab positions in the 2:1 BG1-Env structure (gray; the third Fab is from the model shown in panel C) and the 3:1 BG1-Env structure (green) after superimposing the BG1_A Fabs. (F) Model for binding of two BG1 Fabs to form the 2:1 BG1-Env structure. Panels 1 and 2: BG1_B binds first to the V1V2 epitope on gp120_B such that its CDRL1 interacts with the Asn160gp120 glycan from gp120_A. Panels 3 and 4: BG1_A recognizes the Asn156gp120 and Asn160gp120 glycans from gp120_A. The flexible glycans are shifted away from the trimer 3-fold axis (indicated by arrows in the close-up view of the third panel) such that a potential clash with BG1_B is avoided.